4 Transition to R
Below, we provide some advice and resources if you are transitioning to R.
R was introduced in the late 1990s and has since grown dramatically in scope. Its capabilities are so extensive that commercial alternatives have reacted to R developments in order to stay competitive! (read this article comparing R, SPSS, SAS, STATA, and Python).
Moreover, R is much easier to learn than it was 10 years ago. Previously, R had a reputation of being difficult for beginners. It is now much easier with friendly user-interfaces like RStudio, intuitive code like the tidyverse, and many tutorial resources.
Do not be intimidated - come discover the world of R!

4.1 From Excel
Transitioning from Excel directly to R is a very achievable goal. It may seem daunting, but you can do it!
It is true that someone with strong Excel skills can do very advanced activities in Excel alone - even using scripting tools like VBA. Excel is used across the world and is an essential tool for an epidemiologist. However, complementing it with R can dramatically improve and expand your work flows.
Benefits
You will find that using R offers immense benefits in time saved, more consistent and accurate analysis, reproducibility, shareability, and faster error-correction. Like any new software there is a learning “curve” of time you must invest to become familiar. The dividends will be significant and immense scope of new possibilities will open to you with R.
Excel is a well-known software that can be easy for a beginner to use to produce simple analysis and visualizations with “point-and-click”. In comparison, it can take a couple weeks to become comfortable with R functions and interface. However, R has evolved in recent years to become much more friendly to beginners.
Many Excel workflows rely on memory and on repetition - thus, there is much opportunity for error. Furthermore, generally the data cleaning, analysis methodology, and equations used are hidden from view. It can require substantial time for a new colleague to learn what an Excel workbook is doing and how to troubleshoot it. With R, all the steps are explicitly written in the script and can be easily viewed, edited, corrected, and applied to other datasets.
To begin your transition from Excel to R you must adjust your mindset in a few important ways:
Tidy data
Use machine-readable “tidy” data instead of messy “human-readable” data. These are the three main requirements for “tidy” data, as explained in this tutorial on “tidy” data in R:
- Each variable must have its own column
- Each observation must have its own row
- Each value must have its own cell
To Excel users - think of the role that Excel “tables” play in standardizing data and making the format more predictable.
An example of “tidy” data would be the case linelist used throughout this handbook - each variable is contained within one column, each observation (one case) has it’s own row, and every value is in just one cell. Below you can view the first 50 rows of the linelist:
The main reason one encounters non-tidy data is because many Excel spreadsheets are designed to prioritize easy reading by humans, not easy reading by machines/software.
To help you see the difference, below are some fictional examples of non-tidy data that prioritize human-readability over machine-readability:
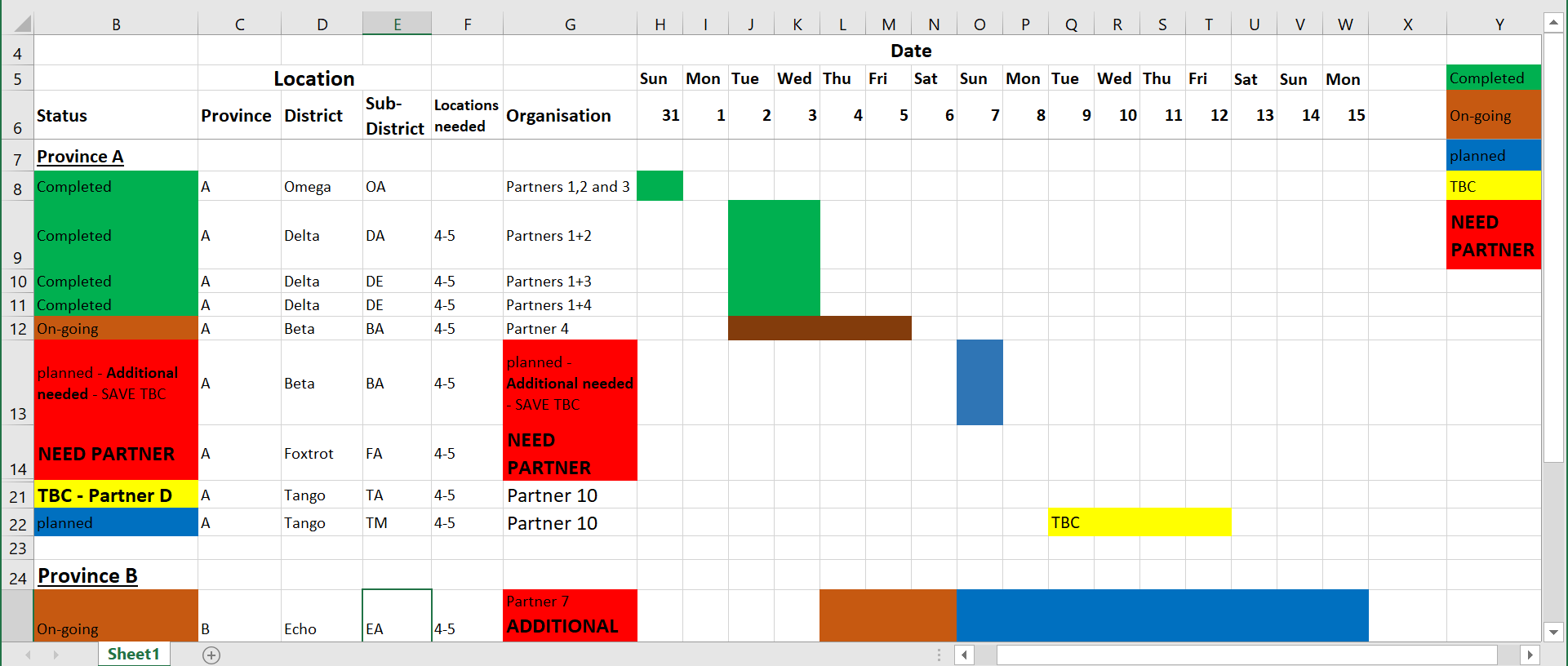
Problems: In the spreadsheet above, there are merged cells which are not easily digested by R. Which row should be considered the “header” is not clear. A color-based dictionary is to the right side and cell values are represented by colors - which is also not easily interpreted by R (nor by humans with color-blindness!). Furthermore, different pieces of information are combined into one cell (multiple partner organizations working in one area, or the status “TBC” in the same cell as “Partner D”).
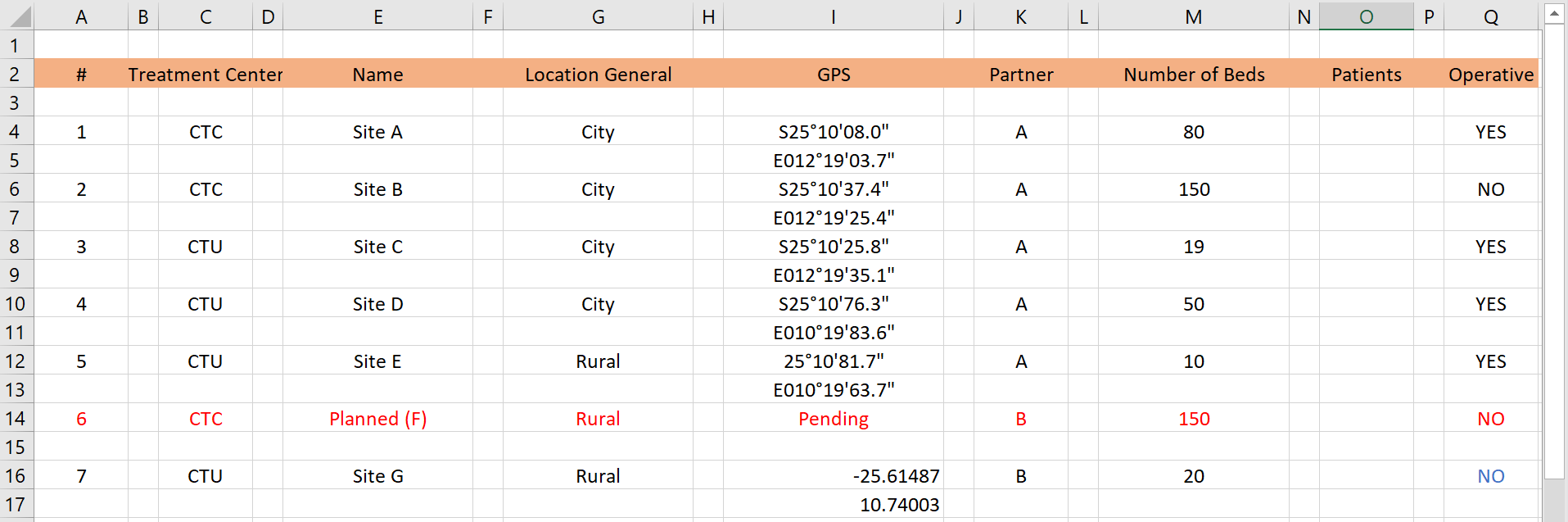
Problems: In the spreadsheet above, there are numerous extra empty rows and columns within the dataset - this will cause cleaning headaches in R. Furthermore, the GPS coordinates are spread across two rows for a given treatment center. As a side note - the GPS coordinates are in two different formats!
“Tidy” datasets may not be as readable to a human eye, but they make data cleaning and analysis much easier! Tidy data can be stored in various formats, for example “long” or “wide”"(see page on Pivoting data), but the principles above are still observed.
Functions
The R word “function” might be new, but the concept exists in Excel too as formulas. Formulas in Excel also require precise syntax (e.g. placement of semicolons and parentheses). All you need to do is learn a few new functions and how they work together in R.
Scripts
Instead of clicking buttons and dragging cells you will be writing every step and procedure into a “script”. Excel users may be familiar with “VBA macros” which also employ a scripting approach.
The R script consists of step-by-step instructions. This allows any colleague to read the script and easily see the steps you took. This also helps de-bug errors or inaccurate calculations. See the [R basics] section on scripts for examples.
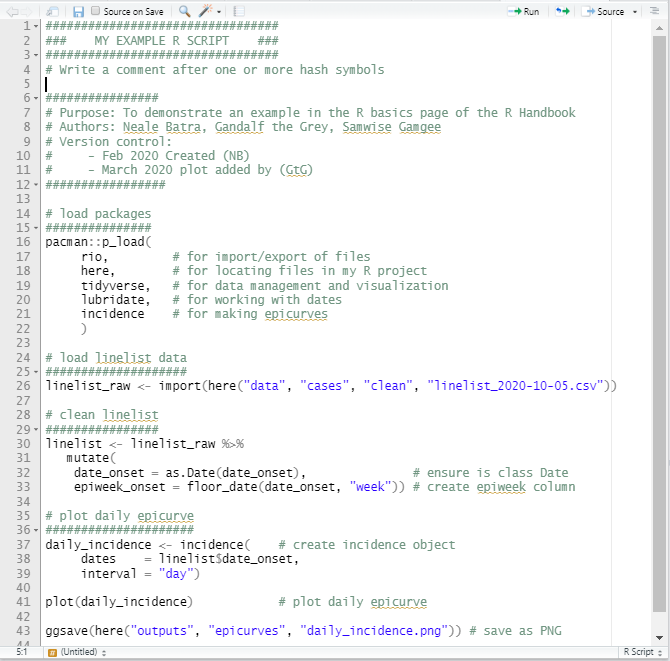
Here is an example of an R script:
R-Excel interaction
R has robust ways to import Excel workbooks, work with the data, export/save Excel files, and work with the nuances of Excel sheets.
It is true that some of the more aesthetic Excel formatting can get lost in translation (e.g. italics, sideways text, etc.). If your work flow requires passing documents back-and-forth between R and Excel while retaining the original Excel formatting, try packages such as openxlsx.
4.2 From Stata
Coming to R from Stata
Many epidemiologists are first taught how to use Stata, and it can seem daunting to move into R. However, if you are a comfortable Stata user then the jump into R is certainly more manageable than you might think. While there are some key differences between Stata and R in how data can be created and modified, as well as how analysis functions are implemented – after learning these key differences you will be able to translate your skills.
Below are some key translations between Stata and R, which may be handy as your review this guide.
General notes
| STATA | R |
|---|---|
| You can only view and manipulate one dataset at a time | You can view and manipulate multiple datasets at the same time, therefore you will frequently have to specify your dataset within the code |
| Online community available through https://www.statalist.org/ | Online community available through RStudio, StackOverFlow, and R-bloggers |
| Point and click functionality as an option | Minimal point and click functionality |
Help for commands available by help [command]
|
Help available by [function]? or search in the Help pane |
| Comment code using * or /// or /* TEXT */ | Comment code using # |
| Almost all commands are built-in to Stata. New/user-written functions can be installed as ado files using ssc install [package] | R installs with base functions, but typical use involves installing other packages from CRAN (see page on [R basics]) |
| Analysis is usually written in a do file | Analysis written in an R script in the RStudio source pane. R markdown scripts are an alternative. |
Working directory
| STATA | R |
|---|---|
| Working directories involve absolute filepaths (e.g. “C:/usename/documents/projects/data/”) | Working directories can be either absolute, or relative to a project root folder by using the here package (see [Import and export]) |
| See current working directory with pwd | Use getwd() or here() (if using the here package), with empty parentheses |
| Set working directory with cd “folder location” | Use setwd(“folder location”), or set_here("folder location) (if using here package) |
Importing and viewing data
| STATA | R |
|---|---|
| Specific commands per file type | Use import() from rio package for almost all filetypes. Specific functions exist as alternatives (see [Import and export]) |
| Reading in csv files is done by import delimited “filename.csv” | Use import("filename.csv")
|
| Reading in xslx files is done by import excel “filename.xlsx” | Use import("filename.xlsx")
|
| Browse your data in a new window using the command browse | View a dataset in the RStudio source pane using View(dataset). You need to specify your dataset name to the function in R because multiple datasets can be held at the same time. Note capital “V” in this function
|
| Get a high-level overview of your dataset using summarize, which provides the variable names and basic information | Get a high-level overview of your dataset using summary(dataset)
|
Basic data manipulation
| STATA | R |
|---|---|
| Dataset columns are often referred to as “variables” | More often referred to as “columns” or sometimes as “vectors” or “variables” |
| No need to specify the dataset | In each of the below commands, you need to specify the dataset - see the page on Cleaning data and core functions for examples |
| New variables are created using the command generate varname = | Generate new variables using the function mutate(varname = ). See page on Cleaning data and core functions for details on all the below dplyr functions. |
| Variables are renamed using rename old_name new_name | Columns can be renamed using the function rename(new_name = old_name)
|
| Variables are dropped using drop varname | Columns can be removed using the function select() with the column name in the parentheses following a minus sign |
| Factor variables can be labeled using a series of commands such as label define | Labeling values can done by converting the column to Factor class and specifying levels. See page on Factors. Column names are not typically labeled as they are in Stata. |
Descriptive analysis
| STATA | R |
|---|---|
| Tabulate counts of a variable using tab varname | Provide the dataset and column name to table() such as table(dataset$colname). Alternatively, use count(varname) from the dplyr package, as explained in Grouping data
|
| Cross-tabulaton of two variables in a 2x2 table is done with tab varname1 varname2 | Use table(dataset$varname1, dataset$varname2 or count(varname1, varname2)
|
While this list gives an overview of the basics in translating Stata commands into R, it is not exhaustive. There are many other great resources for Stata users transitioning to R that could be of interest:
4.3 From SAS
Coming from SAS to R
SAS is commonly used at public health agencies and academic research fields. Although transitioning to a new language is rarely a simple process, understanding key differences between SAS and R may help you start to navigate the new language using your native language. Below outlines the key translations in data management and descriptive analysis between SAS and R.
General notes
| SAS | R |
|---|---|
| Online community available through SAS Customer Support | Online community available through RStudio, StackOverFlow, and R-bloggers |
Help for commands available by help [command]
|
Help available by [function]? or search in the Help pane |
Comment code using * TEXT ; or /* TEXT */
|
Comment code using # |
Almost all commands are built-in. Users can write new functions using SAS macro, SAS/IML, SAS Component Language (SCL), and most recently, procedures Proc Fcmp and Proc Proto
|
R installs with base functions, but typical use involves installing other packages from CRAN (see page on [R basics]) |
| Analysis is usually conducted by writing a SAS program in the Editor window. | Analysis written in an R script in the RStudio source pane. R markdown scripts are an alternative. |
Working directory
| SAS | R |
|---|---|
Working directories can be either absolute, or relative to a project root folder by defining the root folder using %let rootdir=/root path; %include “&rootdir/subfoldername/filename”
|
Working directories can be either absolute, or relative to a project root folder by using the here package (see [Import and export]) |
See current working directory with %put %sysfunc(getoption(work));
|
Use getwd() or here() (if using the here package), with empty parentheses |
Set working directory with libname “folder location”
|
Use setwd(“folder location”), or set_here("folder location) if using here package |
Importing and viewing data
| SAS | R |
|---|---|
Use Proc Import procedure or using Data Step Infile statement. |
Use import() from rio package for almost all filetypes. Specific functions exist as alternatives (see [Import and export]) |
Reading in csv files is done by using Proc Import datafile=”filename.csv” out=work.filename dbms=CSV; run; OR using Data Step Infile statement
|
Use import("filename.csv")
|
Reading in xslx files is done by using Proc Import datafile=”filename.xlsx” out=work.filename dbms=xlsx; run; OR using Data Step Infile statement
|
Use import(“filename.xlsx”) |
| Browse your data in a new window by opening the Explorer window and select desired library and the dataset | View a dataset in the RStudio source pane using View(dataset). You need to specify your dataset name to the function in R because multiple datasets can be held at the same time. Note capital “V” in this function |
Basic data manipulation
| SAS | R |
|---|---|
| Dataset columns are often referred to as “variables” | More often referred to as “columns” or sometimes as “vectors” or “variables” |
| No special procedures are needed to create a variable. New variables are created simply by typing the new variable name, followed by an equal sign, and then an expression for the value | Generate new variables using the function mutate(). See page on Cleaning data and core functions for details on all the below dplyr functions. |
Variables are renamed using rename *old_name=new_name*
|
Columns can be renamed using the function rename(new_name = old_name)
|
Variables are kept using **keep**=varname
|
Columns can be selected using the function select() with the column name in the parentheses |
Variables are dropped using **drop**=varname
|
Columns can be removed using the function select() with the column name in the parentheses following a minus sign |
Factor variables can be labeled in the Data Step using Label statement |
Labeling values can done by converting the column to Factor class and specifying levels. See page on Factors. Column names are not typically labeled. |
Records are selected using Where or If statement in the Data Step. Multiple selection conditions are separated using “and” command. |
Records are selected using the function filter() with multiple selection conditions separated either by an AND operator (&) or a comma |
Datasets are combined using Merge statement in the Data Step. The datasets to be merged need to be sorted first using Proc Sort procedure. |
dplyr package offers a few functions for merging datasets. See page Joining Data for details. |
Descriptive analysis
| SAS | R |
|---|---|
Get a high-level overview of your dataset using Proc Summary procedure, which provides the variable names and descriptive statistics |
Get a high-level overview of your dataset using summary(dataset) or skim(dataset) from the skimr package |
Tabulate counts of a variable using proc freq data=Dataset; Tables varname; Run;
|
See the page on Descriptive tables. Options include table() from base R, and tabyl() from janitor package, among others. Note you will need to specify the dataset and column name as R holds multiple datasets. |
Cross-tabulation of two variables in a 2x2 table is done with proc freq data=Dataset; Tables rowvar*colvar; Run;
|
Again, you can use table(), tabyl() or other options as described in the Descriptive tables page. |
Some useful resources: