41 Organizing routine reports
This page covers the reportfactory package, which is an accompaniment to using R Markdown for reports.
In scenarios where you run reports routinely (daily, weekly, etc.), it eases the compilation of multiple R Markdown files and the organization of their outputs. In essence, it provides a “factory” from which you can run the R Markdown reports, get automatically date- and time-stamped folders for the outputs, and have “light” version control.
reportfactory is one of the packages developed by RECON (R Epidemics Consortium). Here is their website and Github.
41.1 Preparation
Load packages
From within RStudio, install the latest version of the reportfactory package from Github.
You can do this via the pacman package with p_load_current_gh() which will force intall of the latest version from Github. Provide the character string “reconverse/reportfactory”, which specifies the Github organization (reconverse) and repository (reportfactory). You can also use install_github() from the remotes package, as an alternative.
# Install and load the latest version of the package from Github
pacman::p_load_current_gh("reconverse/reportfactory")
#remotes::install_github("reconverse/reportfactory") # alternative41.2 New factory
To create a new factory, run the function new_factory(). This will create a new self-contained R project folder. By default:
- The factory will be added to your working directory
- The name of the factory R project will be called “new_factory.Rproj”
- Your RStudio session will “move in” to this R project
# This will create the factory in the working directory
new_factory()Looking inside the factory, you can see that sub-folders and some files were created automatically.
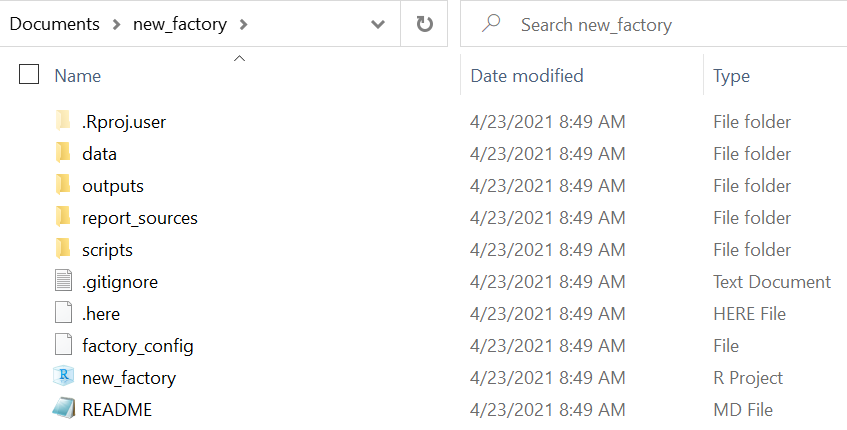
- The report_sources folder will hold your R Markdown scripts, which generate your reports
- The outputs folder will hold the report outputs (e.g. HTML, Word, PDF, etc.)
- The scripts folder can be used to store other R scripts (e.g. that are sourced by your Rmd scripts)
- The data folder can be used to hold your data (“raw” and “clean” subfolders are included)
- A .here file, so you can use the here package to call files in sub-folders by their relation to this root folder (see [R projects] page for details)
- A gitignore file was created in case you link this R project to a Github repository (see [Version control and collaboration with Github])
- An empty README file, for if you use a Github repository
CAUTION: depending on your computer’s setting, files such as “.here” may exist but be invisible.
Of the default settings, below are several that you might want to adjust within the new_factory() command:
-
factory =- Provide a name for the factory folder (default is “new_factory”)
-
path =- Designate a file path for the new factory (default is the working directory)
-
report_sources =Provide an alternate name for the subfolder which holds the R Markdown scripts (default is “report_sources”)
-
outputs =Provide an alternate name for the folder which holds the report outputs (default is “outputs”)
See ?new_factory for a complete list of the arguments.
When you create the new factory, your R session is transferred to the new R project, so you should again load the reportfactory package.
pacman::p_load(reportfactory)Now you can run a the factory_overview() command to see the internal structure (all folders and files) in the factory.
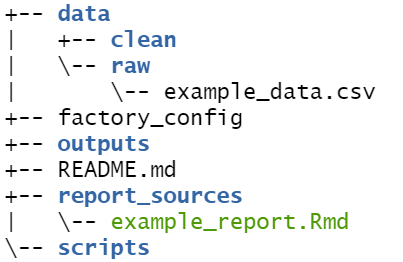
factory_overview() # print overview of the factory to consoleThe following “tree” of the factory’s folders and files is printed to the R console. Note that in the “data” folder there are sub-folders for “raw” and “clean” data, and example CSV data. There is also “example_report.Rmd” in the “report_sources” folder.
41.3 Create a report
From within the factory R project, create a R Markdown report just as you would normally, and save it into the “report_sources” folder. See the R Markdown page for instructions. For purposes of example, we have added the following to the factory:
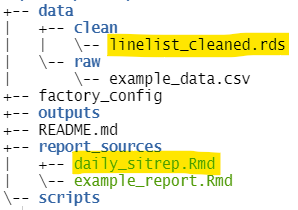
- A new R markdown script entitled “daily_sitrep.Rmd”, saved within the “report_sources” folder
- Data for the report (“linelist_cleaned.rds”), saved to the “clean” sub-folder within the “data” folder
We can see using factory_overview() our R Markdown in the “report_sources” folder and the data file in the “clean” data folder (highlighted):
Below is a screenshot of the beginning of the R Markdown “daily_sitrep.Rmd”. You can see that the output format is set to be HTML, via the YAML header output: html_document.
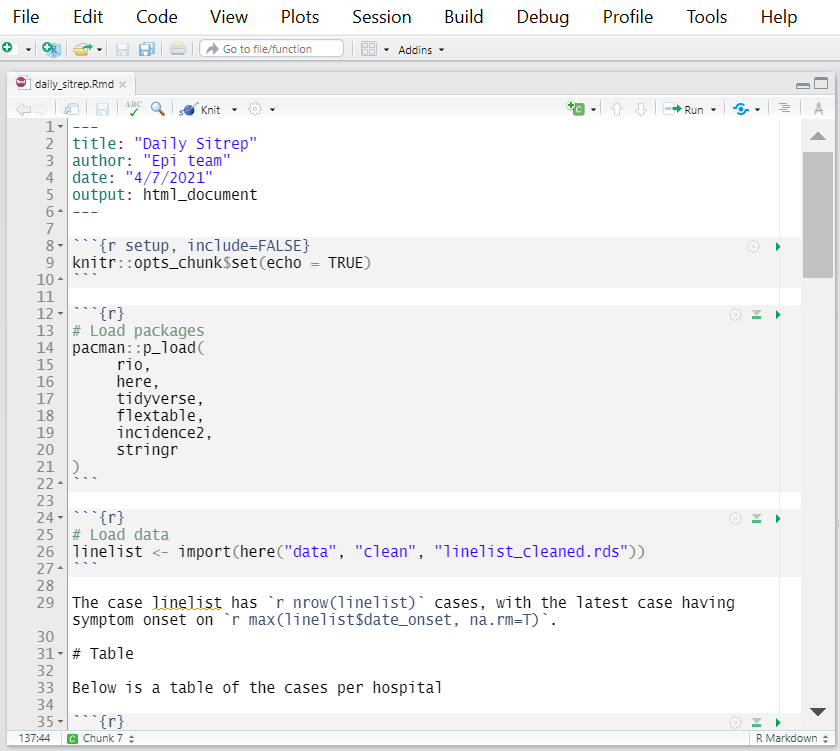
In this simple script, there are commands to:
- Load necessary packages
- Import the linelist data using a filepath from the here package (read more in the page on [Import and export])
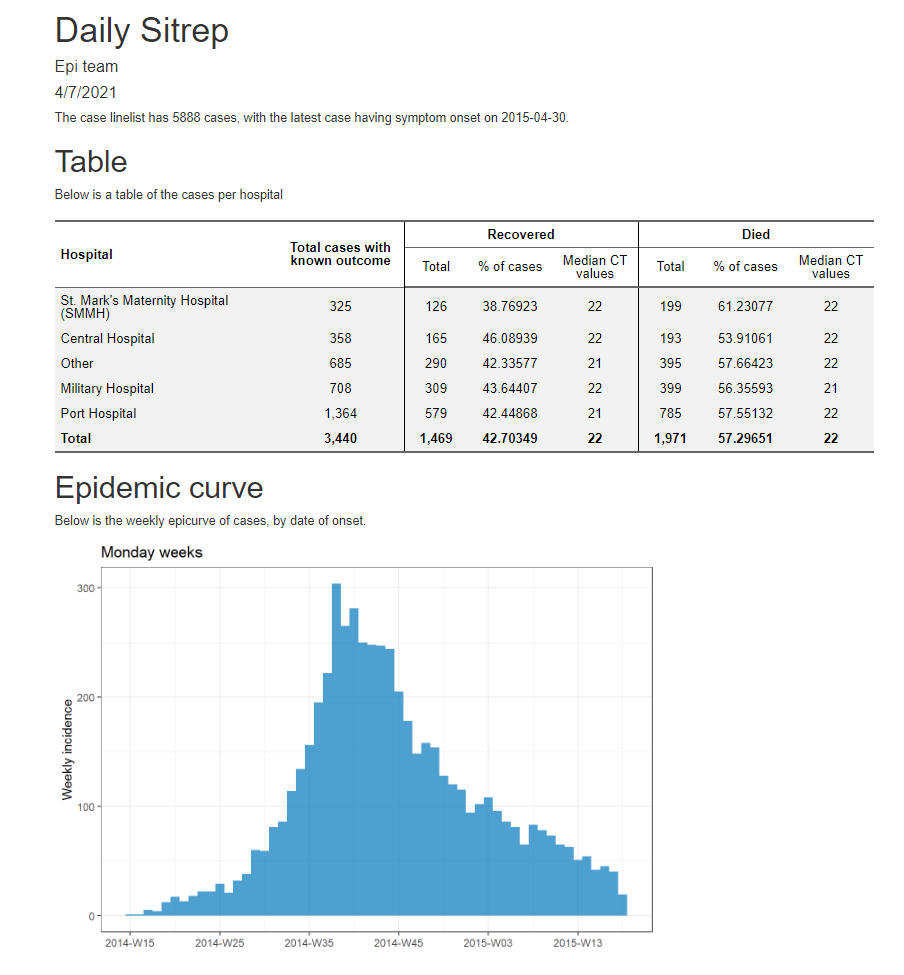
- Print a summary table of cases, and export it with
export()as a .csv file
- Print an epicurve, and export it with
ggsave()as a .png file
You can review just the list of R Markdown reports in the “report_sources” folder with this command:
list_reports()41.4 Compile
In a report factory, to “compile” a R Markdown report means that the .Rmd script will be run and the output will be produced (as specified in the script YAML e.g. as HTML, Word, PDF, etc).
The factory will automatically create a date- and time-stamped folder for the outputs in the “outputs” folder.
The report itself and any exported files produced by the script (e.g. csv, png, xlsx) will be saved into this folder. In addition, the Rmd script itself will be saved in this folder, so you have a record of that version of the script.
This contrasts with the normal behavior of a “knitted” R Markdown, which saves outputs to the location of the Rmd script. This default behavior can result in crowded, messy folders. The factory aims to improve organization when one needs to run reports frequently.
Compile by name
You can compile a specific report by running compile_reports() and providing the Rmd script name (without .Rmd extension) to reports =. For simplicity, you can skip the reports = and just write the R Markdown name in quotes, as below.

This command would compile only the “daily_sitrep.Rmd” report, saving the HTML report, and the .csv table and .png epicurve exports into a date- and time-stamped sub-folder specific to the report, within the “outputs” folder.
Note that if you choose to provide the .Rmd extension, you must correctly type the extension as it is saved in the file name (.rmd vs. .Rmd).
Also note that when you compile, you may see several files temporarily appear in the “report_sources” folder - but they will soon disappear as they are transferred to the correct “outputs” folder.
Compile by number
You can also specify the Rmd script to compile by providing a number or vector of numbers to reports =. The numbers must align with the order the reports appear when you run list_reports().
# Compile the second and fourth Rmds in the "report_sources" folder
compile_reports(reports = c(2, 4))Compile all
You can compile all the R Markdown reports in the “report_sources” folder by setting the reports = argument to TRUE.

Compile from sub-folder
You can add sub-folders to the “report_sources” folder. To run an R Markdown report from a subfolder, simply provide the name of the folder to subfolder =. Below is an example of code to compile a Rmd report that lives in a sub_folder of “report_sources”.
compile_reports(
reports = "summary_for_partners.Rmd",
subfolder = "for_partners")You can compile all Rmd reports within a subfolder by providing the subfolder name to reports =, with a slash on the end, as below.
compile_reports(reports = "for_partners/")Parameterization
As noted in the page on Reports with R Markdown, you can run reports with specified parameters. You can pass these parameters as a list to compile_reports() via the params = argument. For example, in this fictional report there are three parameters provided to the R Markdown reports.
compile_reports(
reports = "daily_sitrep.Rmd",
params = list(most_recent_data = TRUE,
region = "NORTHERN",
rates_denominator = 10000),
subfolder = "regional"
)41.5 Outputs
After we have compiled the reports a few times, the “outputs” folder might look like this (highlights added for clarity):
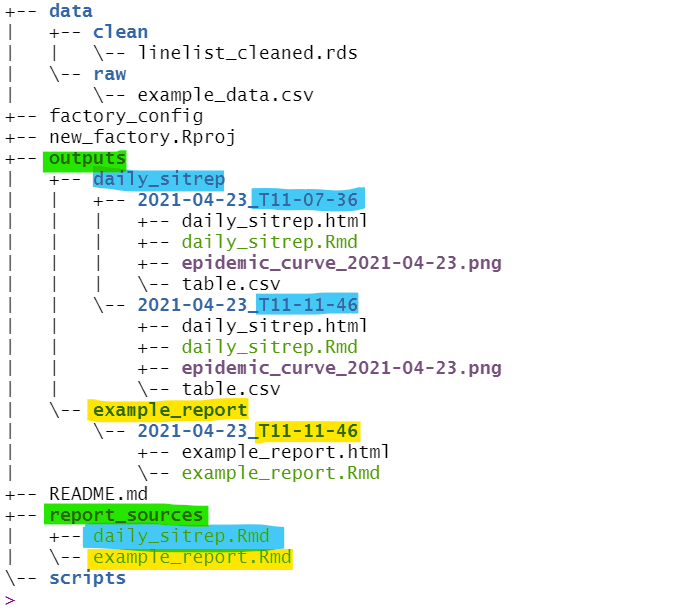
- Within “outputs”, sub-folders have been created for each Rmd report
- Within those, further sub-folders have been created for each unique compiling
- These are date- and time-stamped (“2021-04-23_T11-07-36” means 23rd April 2021 at 11:07:36)
- You can edit the date/time-stamp format. See
?compile_reports
- These are date- and time-stamped (“2021-04-23_T11-07-36” means 23rd April 2021 at 11:07:36)
- Within each date/time compiled folder, the report output is stored (e.g. HTML, PDF, Word) along with the Rmd script (version control!) and any other exported files (e.g. table.csv, epidemic_curve.png)
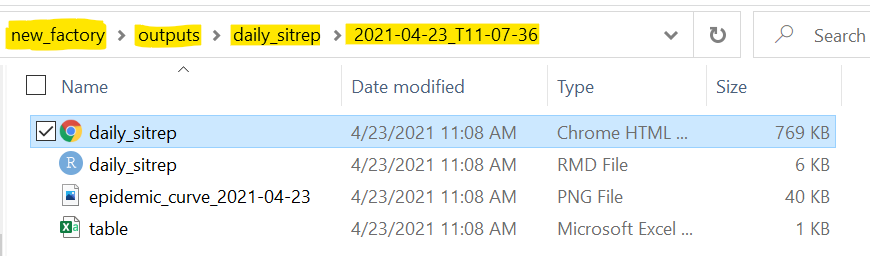
Here is a view inside one of the date/time-stamped folders, for the “daily_sitrep” report. The file path is highlighted in yellow for emphasis.
Finally, below is a screenshot of the HTML report output.
You can use list_outputs() to review a list of the outputs.
41.6 Miscellaneous
Knit
You can still “knit” one of your R Markdown reports by pressing the “Knit” button, if you want. If you do this, as by default, the outputs will appear in the folder where the Rmd is saved - the “report_sources” folder. In prior versions of reportfactory, having any non-Rmd files in “report_sources” would prevent compiling, but this is no longer the case. You can run compile_reports() and no error will occur.
Scripts
We encourage you to utilize the “scripts” folder to store “runfiles” or .R scripts that are sourced by your .Rmd scripts. See the page on R Markdown for tips on how to structure your code across several files.
Extras
With reportfactory, you can use the function
list_deps()to list all packages required across all the reports in the entire factory.-
There is an accompanying package in development called rfextras that offers more helper functions to assist you in building reports, such as:
-
load_scripts()- sources/loads all .R scripts in a given folder (the “scripts” folder by default)
-
find_latest()- finds the latest version of a file (e.g. the latest dataset)
-