8 Cleaning data and core functions
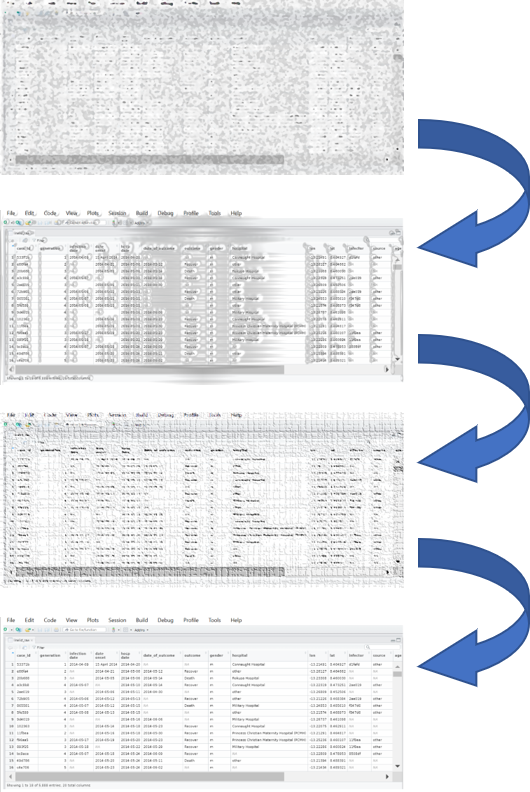
This page demonstrates common steps used in the process of “cleaning” a dataset, and also explains the use of many essential R data management functions.
To demonstrate data cleaning, this page begins by importing a raw case linelist dataset, and proceeds step-by-step through the cleaning process. In the R code, this manifests as a “pipe” chain, which references the “pipe” operator %>% that passes a dataset from one operation to the next.
Core functions
This handbook emphasizes use of the functions from the tidyverse family of R packages. The essential R functions demonstrated in this page are listed below.
Many of these functions belong to the dplyr R package, which provides “verb” functions to solve data manipulation challenges (the name is a reference to a "data frame-plier. dplyr is part of the tidyverse family of R packages (which also includes ggplot2, tidyr, stringr, tibble, purrr, magrittr, and forcats among others).
| Function | Utility | Package |
|---|---|---|
%>% |
“pipe” (pass) data from one function to the next | magrittr |
mutate() |
create, transform, and re-define columns | dplyr |
select() |
keep, remove, select, or re-name columns | dplyr |
rename() |
rename columns | dplyr |
clean_names() |
standardize the syntax of column names | janitor |
as.character(), as.numeric(), as.Date(), etc. |
convert the class of a column | base R |
across() |
transform multiple columns at one time | dplyr |
| tidyselect functions | use logic to select columns | tidyselect |
filter() |
keep certain rows | dplyr |
distinct() |
de-duplicate rows | dplyr |
rowwise() |
operations by/within each row | dplyr |
add_row() |
add rows manually | tibble |
arrange() |
sort rows | dplyr |
recode() |
re-code values in a column | dplyr |
case_when() |
re-code values in a column using more complex logical criteria | dplyr |
replace_na(), na_if(), coalesce()
|
special functions for re-coding | tidyr |
age_categories() and cut()
|
create categorical groups from a numeric column | epikit and base R |
clean_variable_spelling() |
re-code/clean values using a data dictionary | linelist |
which() |
apply logical criteria; return indices | base R |
If you want to see how these functions compare to Stata or SAS commands, see the page on Transition to R.
You may encounter an alternative data management framework from the data.table R package with operators like := and frequent use of brackets [ ]. This approach and syntax is briefly explained in the Data Table page.
Nomenclature
In this handbook, we generally reference “columns” and “rows” instead of “variables” and “observations”. As explained in this primer on “tidy data”, most epidemiological statistical datasets consist structurally of rows, columns, and values.
Variables contain the values that measure the same underlying attribute (like age group, outcome, or date of onset). Observations contain all values measured on the same unit (e.g. a person, site, or lab sample). So these aspects can be more difficult to tangibly define.
In “tidy” datasets, each column is a variable, each row is an observation, and each cell is a single value. However some datasets you encounter will not fit this mold - a “wide” format dataset may have a variable split across several columns (see an example in the Pivoting data page). Likewise, observations could be split across several rows.
Most of this handbook is about managing and transforming data, so referring to the concrete data structures of rows and columns is more relevant than the more abstract observations and variables. Exceptions occur primarily in pages on data analysis, where you will see more references to variables and observations.
8.1 Cleaning pipeline
This page proceeds through typical cleaning steps, adding them sequentially to a cleaning pipe chain.
In epidemiological analysis and data processing, cleaning steps are often performed sequentially, linked together. In R, this often manifests as a cleaning “pipeline”, where the raw dataset is passed or “piped” from one cleaning step to another.
Such chains utilize dplyr “verb” functions and the magrittr pipe operator %>%. This pipe begins with the “raw” data (“linelist_raw.xlsx”) and ends with a “clean” R data frame (linelist) that can be used, saved, exported, etc.
In a cleaning pipeline the order of the steps is important. Cleaning steps might include:
- Importing of data
- Column names cleaned or changed
- De-duplication
- Column creation and transformation (e.g. re-coding or standardising values)
- Rows filtered or added
8.2 Load packages
This code chunk shows the loading of packages required for the analyses. In this handbook we emphasize p_load() from pacman, which installs the package if necessary and loads it for use. You can also load installed packages with library() from base R. See the page on [R basics] for more information on R packages.
pacman::p_load(
rio, # importing data
here, # relative file pathways
janitor, # data cleaning and tables
lubridate, # working with dates
epikit, # age_categories() function
tidyverse # data management and visualization
)8.3 Import data
Import
Here we import the “raw” case linelist Excel file using the import() function from the package rio. The rio package flexibly handles many types of files (e.g. .xlsx, .csv, .tsv, .rds. See the page on [Import and export] for more information and tips on unusual situations (e.g. skipping rows, setting missing values, importing Google sheets, etc).
If you want to follow along, click to download the “raw” linelist (as .xlsx file).
If your dataset is large and takes a long time to import, it can be useful to have the import command be separate from the pipe chain and the “raw” saved as a distinct file. This also allows easy comparison between the original and cleaned versions.
Below we import the raw Excel file and save it as the data frame linelist_raw. We assume the file is located in your working directory or R project root, and so no sub-folders are specified in the file path.
linelist_raw <- import("linelist_raw.xlsx")You can view the first 50 rows of the the data frame below. Note: the base R function head(n) allow you to view just the first n rows in the R console.
Review
You can use the function skim() from the package skimr to get an overview of the entire dataframe (see page on Descriptive tables for more info). Columns are summarised by class/type such as character, numeric. Note: “POSIXct” is a type of raw date class (see Working with dates.
skimr::skim(linelist_raw)| Name | linelist_raw |
| Number of rows | 6611 |
| Number of columns | 28 |
| _______________________ | |
| Column type frequency: | |
| character | 17 |
| numeric | 8 |
| POSIXct | 3 |
| ________________________ | |
| Group variables | None |
Variable type: character
| skim_variable | n_missing | complete_rate | min | max | empty | n_unique | whitespace |
|---|---|---|---|---|---|---|---|
| case_id | 137 | 0.98 | 6 | 6 | 0 | 5888 | 0 |
| date onset | 293 | 0.96 | 10 | 10 | 0 | 580 | 0 |
| outcome | 1500 | 0.77 | 5 | 7 | 0 | 2 | 0 |
| gender | 324 | 0.95 | 1 | 1 | 0 | 2 | 0 |
| hospital | 1512 | 0.77 | 5 | 36 | 0 | 13 | 0 |
| infector | 2323 | 0.65 | 6 | 6 | 0 | 2697 | 0 |
| source | 2323 | 0.65 | 5 | 7 | 0 | 2 | 0 |
| age | 107 | 0.98 | 1 | 2 | 0 | 75 | 0 |
| age_unit | 7 | 1.00 | 5 | 6 | 0 | 2 | 0 |
| fever | 258 | 0.96 | 2 | 3 | 0 | 2 | 0 |
| chills | 258 | 0.96 | 2 | 3 | 0 | 2 | 0 |
| cough | 258 | 0.96 | 2 | 3 | 0 | 2 | 0 |
| aches | 258 | 0.96 | 2 | 3 | 0 | 2 | 0 |
| vomit | 258 | 0.96 | 2 | 3 | 0 | 2 | 0 |
| time_admission | 844 | 0.87 | 5 | 5 | 0 | 1091 | 0 |
| merged_header | 0 | 1.00 | 1 | 1 | 0 | 1 | 0 |
| …28 | 0 | 1.00 | 1 | 1 | 0 | 1 | 0 |
Variable type: numeric
| skim_variable | n_missing | complete_rate | mean | sd | p0 | p25 | p50 | p75 | p100 |
|---|---|---|---|---|---|---|---|---|---|
| generation | 7 | 1.00 | 16.60 | 5.71 | 0.00 | 13.00 | 16.00 | 20.00 | 37.00 |
| lon | 7 | 1.00 | -13.23 | 0.02 | -13.27 | -13.25 | -13.23 | -13.22 | -13.21 |
| lat | 7 | 1.00 | 8.47 | 0.01 | 8.45 | 8.46 | 8.47 | 8.48 | 8.49 |
| row_num | 0 | 1.00 | 3240.91 | 1857.83 | 1.00 | 1647.50 | 3241.00 | 4836.50 | 6481.00 |
| wt_kg | 7 | 1.00 | 52.69 | 18.59 | -11.00 | 41.00 | 54.00 | 66.00 | 111.00 |
| ht_cm | 7 | 1.00 | 125.25 | 49.57 | 4.00 | 91.00 | 130.00 | 159.00 | 295.00 |
| ct_blood | 7 | 1.00 | 21.26 | 1.67 | 16.00 | 20.00 | 22.00 | 22.00 | 26.00 |
| temp | 158 | 0.98 | 38.60 | 0.95 | 35.20 | 38.30 | 38.80 | 39.20 | 40.80 |
Variable type: POSIXct
| skim_variable | n_missing | complete_rate | min | max | median | n_unique |
|---|---|---|---|---|---|---|
| infection date | 2322 | 0.65 | 2012-04-09 | 2015-04-27 | 2014-10-04 | 538 |
| hosp date | 7 | 1.00 | 2012-04-20 | 2015-04-30 | 2014-10-15 | 570 |
| date_of_outcome | 1068 | 0.84 | 2012-05-14 | 2015-06-04 | 2014-10-26 | 575 |
8.4 Column names
In R, column names are the “header” or “top” value of a column. They are used to refer to columns in the code, and serve as a default label in figures.
Other statistical software such as SAS and STATA use “labels” that co-exist as longer printed versions of the shorter column names. While R does offer the possibility of adding column labels to the data, this is not emphasized in most practice. To make column names “printer-friendly” for figures, one typically adjusts their display within the plotting commands that create the outputs (e.g. axis or legend titles of a plot, or column headers in a printed table - see the scales section of the ggplot tips page and Tables for presentation pages). If you want to assign column labels in the data, read more online here and here.
As R column names are used very often, so they must have “clean” syntax. We suggest the following:
- Short names
- No spaces (replace with underscores _ )
- No unusual characters (&, #, <, >, …)
- Similar style nomenclature (e.g. all date columns named like date_onset, date_report, date_death…)
The columns names of linelist_raw are printed below using names() from base R. We can see that initially:
- Some names contain spaces (e.g.
infection date)
- Different naming patterns are used for dates (
date onsetvs.infection date)
- There must have been a merged header across the two last columns in the .xlsx. We know this because the name of two merged columns (“merged_header”) was assigned by R to the first column, and the second column was assigned a placeholder name “…28” (as it was then empty and is the 28th column).
names(linelist_raw)## [1] "case_id" "generation" "infection date" "date onset" "hosp date" "date_of_outcome"
## [7] "outcome" "gender" "hospital" "lon" "lat" "infector"
## [13] "source" "age" "age_unit" "row_num" "wt_kg" "ht_cm"
## [19] "ct_blood" "fever" "chills" "cough" "aches" "vomit"
## [25] "temp" "time_admission" "merged_header" "...28"NOTE: To reference a column name that includes spaces, surround the name with back-ticks, for example: linelist$` '\x60infection date\x60'`. note that on your keyboard, the back-tick (`) is different from the single quotation mark (’).
Automatic cleaning
The function clean_names() from the package janitor standardizes column names and makes them unique by doing the following:
- Converts all names to consist of only underscores, numbers, and letters
- Accented characters are transliterated to ASCII (e.g. german o with umlaut becomes “o”, spanish “enye” becomes “n”)
- Capitalization preference for the new column names can be specified using the
case =argument (“snake” is default, alternatives include “sentence”, “title”, “small_camel”…)
- You can specify specific name replacements by providing a vector to the
replace =argument (e.g.replace = c(onset = "date_of_onset"))
- Here is an online vignette
Below, the cleaning pipeline begins by using clean_names() on the raw linelist.
# pipe the raw dataset through the function clean_names(), assign result as "linelist"
linelist <- linelist_raw %>%
janitor::clean_names()
# see the new column names
names(linelist)## [1] "case_id" "generation" "infection_date" "date_onset" "hosp_date" "date_of_outcome"
## [7] "outcome" "gender" "hospital" "lon" "lat" "infector"
## [13] "source" "age" "age_unit" "row_num" "wt_kg" "ht_cm"
## [19] "ct_blood" "fever" "chills" "cough" "aches" "vomit"
## [25] "temp" "time_admission" "merged_header" "x28"NOTE: The last column name “…28” was changed to “x28”.
Manual name cleaning
Re-naming columns manually is often necessary, even after the standardization step above. Below, re-naming is performed using the rename() function from the dplyr package, as part of a pipe chain. rename() uses the style NEW = OLD - the new column name is given before the old column name.
Below, a re-naming command is added to the cleaning pipeline. Spaces have been added strategically to align code for easier reading.
# CLEANING 'PIPE' CHAIN (starts with raw data and pipes it through cleaning steps)
##################################################################################
linelist <- linelist_raw %>%
# standardize column name syntax
janitor::clean_names() %>%
# manually re-name columns
# NEW name # OLD name
rename(date_infection = infection_date,
date_hospitalisation = hosp_date,
date_outcome = date_of_outcome)Now you can see that the columns names have been changed:
## [1] "case_id" "generation" "date_infection" "date_onset" "date_hospitalisation"
## [6] "date_outcome" "outcome" "gender" "hospital" "lon"
## [11] "lat" "infector" "source" "age" "age_unit"
## [16] "row_num" "wt_kg" "ht_cm" "ct_blood" "fever"
## [21] "chills" "cough" "aches" "vomit" "temp"
## [26] "time_admission" "merged_header" "x28"Rename by column position
You can also rename by column position, instead of column name, for example:
rename(newNameForFirstColumn = 1,
newNameForSecondColumn = 2)Rename via select() and summarise()
As a shortcut, you can also rename columns within the dplyr select() and summarise() functions. select() is used to keep only certain columns (and is covered later in this page). summarise() is covered in the Grouping data and Descriptive tables pages. These functions also uses the format new_name = old_name. Here is an example:
linelist_raw %>%
select(# NEW name # OLD name
date_infection = `infection date`, # rename and KEEP ONLY these columns
date_hospitalisation = `hosp date`)Other challenges
Empty Excel column names
R cannot have dataset columns that do not have column names (headers). So, if you import an Excel dataset with data but no column headers, R will fill-in the headers with names like “…1” or “…2”. The number represents the column number (e.g. if the 4th column in the dataset has no header, then R will name it “…4”).
You can clean these names manually by referencing their position number (see example above), or their assigned name (linelist_raw$...1).
Merged Excel column names and cells
Merged cells in an Excel file are a common occurrence when receiving data. As explained in Transition to R, merged cells can be nice for human reading of data, but are not “tidy data” and cause many problems for machine reading of data. R cannot accommodate merged cells.
Remind people doing data entry that human-readable data is not the same as machine-readable data. Strive to train users about the principles of tidy data. If at all possible, try to change procedures so that data arrive in a tidy format without merged cells.
- Each variable must have its own column.
- Each observation must have its own row.
- Each value must have its own cell.
When using rio’s import() function, the value in a merged cell will be assigned to the first cell and subsequent cells will be empty.
One solution to deal with merged cells is to import the data with the function readWorkbook() from the package openxlsx. Set the argument fillMergedCells = TRUE. This gives the value in a merged cell to all cells within the merge range.
linelist_raw <- openxlsx::readWorkbook("linelist_raw.xlsx", fillMergedCells = TRUE)DANGER: If column names are merged with readWorkbook(), you will end up with duplicate column names, which you will need to fix manually - R does not work well with duplicate column names! You can re-name them by referencing their position (e.g. column 5), as explained in the section on manual column name cleaning.
8.5 Select or re-order columns
Use select() from dplyr to select the columns you want to retain, and to specify their order in the data frame.
CAUTION: In the examples below, the linelist data frame is modified with select() and displayed, but not saved. This is for demonstration purposes. The modified column names are printed by piping the data frame to names().
Here are ALL the column names in the linelist at this point in the cleaning pipe chain:
names(linelist)## [1] "case_id" "generation" "date_infection" "date_onset" "date_hospitalisation"
## [6] "date_outcome" "outcome" "gender" "hospital" "lon"
## [11] "lat" "infector" "source" "age" "age_unit"
## [16] "row_num" "wt_kg" "ht_cm" "ct_blood" "fever"
## [21] "chills" "cough" "aches" "vomit" "temp"
## [26] "time_admission" "merged_header" "x28"Keep columns
Select only the columns you want to remain
Put their names in the select() command, with no quotation marks. They will appear in the data frame in the order you provide. Note that if you include a column that does not exist, R will return an error (see use of any_of() below if you want no error in this situation).
# linelist dataset is piped through select() command, and names() prints just the column names
linelist %>%
select(case_id, date_onset, date_hospitalisation, fever) %>%
names() # display the column names## [1] "case_id" "date_onset" "date_hospitalisation" "fever"“tidyselect” helper functions
These helper functions exist to make it easy to specify columns to keep, discard, or transform. They are from the package tidyselect, which is included in tidyverse and underlies how columns are selected in dplyr functions.
For example, if you want to re-order the columns, everything() is a useful function to signify “all other columns not yet mentioned”. The command below moves columns date_onset and date_hospitalisation to the beginning (left) of the dataset, but keeps all the other columns afterward. Note that everything() is written with empty parentheses:
# move date_onset and date_hospitalisation to beginning
linelist %>%
select(date_onset, date_hospitalisation, everything()) %>%
names()## [1] "date_onset" "date_hospitalisation" "case_id" "generation" "date_infection"
## [6] "date_outcome" "outcome" "gender" "hospital" "lon"
## [11] "lat" "infector" "source" "age" "age_unit"
## [16] "row_num" "wt_kg" "ht_cm" "ct_blood" "fever"
## [21] "chills" "cough" "aches" "vomit" "temp"
## [26] "time_admission" "merged_header" "x28"Here are other “tidyselect” helper functions that also work within dplyr functions like select(), across(), and summarise():
everything()- all other columns not mentionedlast_col()- the last columnwhere()- applies a function to all columns and selects those which are TRUE-
contains()- columns containing a character string- example:
select(contains("time"))
- example:
-
starts_with()- matches to a specified prefix- example:
select(starts_with("date_"))
- example:
-
ends_with()- matches to a specified suffix- example:
select(ends_with("_post"))
- example:
-
matches()- to apply a regular expression (regex)- example:
select(matches("[pt]al"))
- example:
num_range()- a numerical range like x01, x02, x03-
any_of()- matches IF column exists but returns no error if it is not found- example:
select(any_of(date_onset, date_death, cardiac_arrest))
- example:
In addition, use normal operators such as c() to list several columns, : for consecutive columns, ! for opposite, & for AND, and | for OR.
Use where() to specify logical criteria for columns. If providing a function inside where(), do not include the function’s empty parentheses. The command below selects columns that are class Numeric.
# select columns that are class Numeric
linelist %>%
select(where(is.numeric)) %>%
names()## [1] "generation" "lon" "lat" "row_num" "wt_kg" "ht_cm" "ct_blood" "temp"Use contains() to select only columns in which the column name contains a specified character string. ends_with() and starts_with() provide more nuance.
# select columns containing certain characters
linelist %>%
select(contains("date")) %>%
names()## [1] "date_infection" "date_onset" "date_hospitalisation" "date_outcome"The function matches() works similarly to contains() but can be provided a regular expression (see page on Characters and strings), such as multiple strings separated by OR bars within the parentheses:
# searched for multiple character matches
linelist %>%
select(matches("onset|hosp|fev")) %>% # note the OR symbol "|"
names()## [1] "date_onset" "date_hospitalisation" "hospital" "fever"CAUTION: If a column name that you specifically provide does not exist in the data, it can return an error and stop your code. Consider using any_of() to cite columns that may or may not exist, especially useful in negative (remove) selections.
Only one of these columns exists, but no error is produced and the code continues without stopping your cleaning chain.
linelist %>%
select(any_of(c("date_onset", "village_origin", "village_detection", "village_residence", "village_travel"))) %>%
names()## [1] "date_onset"Remove columns
Indicate which columns to remove by placing a minus symbol “-” in front of the column name (e.g. select(-outcome)), or a vector of column names (as below). All other columns will be retained.
linelist %>%
select(-c(date_onset, fever:vomit)) %>% # remove date_onset and all columns from fever to vomit
names()## [1] "case_id" "generation" "date_infection" "date_hospitalisation" "date_outcome"
## [6] "outcome" "gender" "hospital" "lon" "lat"
## [11] "infector" "source" "age" "age_unit" "row_num"
## [16] "wt_kg" "ht_cm" "ct_blood" "temp" "time_admission"
## [21] "merged_header" "x28"You can also remove a column using base R syntax, by defining it as NULL. For example:
linelist$date_onset <- NULL # deletes column with base R syntax Standalone
select() can also be used as an independent command (not in a pipe chain). In this case, the first argument is the original dataframe to be operated upon.
# Create a new linelist with id and age-related columns
linelist_age <- select(linelist, case_id, contains("age"))
# display the column names
names(linelist_age)## [1] "case_id" "age" "age_unit"Add to the pipe chain
In the linelist_raw, there are a few columns we do not need: row_num, merged_header, and x28. We remove them with a select() command in the cleaning pipe chain:
# CLEANING 'PIPE' CHAIN (starts with raw data and pipes it through cleaning steps)
##################################################################################
# begin cleaning pipe chain
###########################
linelist <- linelist_raw %>%
# standardize column name syntax
janitor::clean_names() %>%
# manually re-name columns
# NEW name # OLD name
rename(date_infection = infection_date,
date_hospitalisation = hosp_date,
date_outcome = date_of_outcome) %>%
# ABOVE ARE UPSTREAM CLEANING STEPS ALREADY DISCUSSED
#####################################################
# remove column
select(-c(row_num, merged_header, x28))8.6 Deduplication
See the handbook page on De-duplication for extensive options on how to de-duplicate data. Only a very simple row de-duplication example is presented here.
The package dplyr offers the distinct() function. This function examines every row and reduce the data frame to only the unique rows. That is, it removes rows that are 100% duplicates.
When evaluating duplicate rows, it takes into account a range of columns - by default it considers all columns. As shown in the de-duplication page, you can adjust this column range so that the uniqueness of rows is only evaluated in regards to certain columns.
In this simple example, we just add the empty command distinct() to the pipe chain. This ensures there are no rows that are 100% duplicates of other rows (evaluated across all columns).
We begin with nrow(linelist) rows in linelist.
linelist <- linelist %>%
distinct()After de-duplication there are nrow(linelist) rows. Any removed rows would have been 100% duplicates of other rows.
Below, the distinct() command is added to the cleaning pipe chain:
# CLEANING 'PIPE' CHAIN (starts with raw data and pipes it through cleaning steps)
##################################################################################
# begin cleaning pipe chain
###########################
linelist <- linelist_raw %>%
# standardize column name syntax
janitor::clean_names() %>%
# manually re-name columns
# NEW name # OLD name
rename(date_infection = infection_date,
date_hospitalisation = hosp_date,
date_outcome = date_of_outcome) %>%
# remove column
select(-c(row_num, merged_header, x28)) %>%
# ABOVE ARE UPSTREAM CLEANING STEPS ALREADY DISCUSSED
#####################################################
# de-duplicate
distinct()8.7 Column creation and transformation
We recommend using the dplyr function mutate() to add a new column, or to modify an existing one.
Below is an example of creating a new column with mutate(). The syntax is: mutate(new_column_name = value or transformation)
In Stata, this is similar to the command generate, but R’s mutate() can also be used to modify an existing column.
New columns
The most basic mutate() command to create a new column might look like this. It creates a new column new_col where the value in every row is 10.
linelist <- linelist %>%
mutate(new_col = 10)You can also reference values in other columns, to perform calculations. Below, a new column bmi is created to hold the Body Mass Index (BMI) for each case - as calculated using the formula BMI = kg/m^2, using column ht_cm and column wt_kg.
linelist <- linelist %>%
mutate(bmi = wt_kg / (ht_cm/100)^2)If creating multiple new columns, separate each with a comma and new line. Below are examples of new columns, including ones that consist of values from other columns combined using str_glue() from the stringr package (see page on Characters and strings.
new_col_demo <- linelist %>%
mutate(
new_var_dup = case_id, # new column = duplicate/copy another existing column
new_var_static = 7, # new column = all values the same
new_var_static = new_var_static + 5, # you can overwrite a column, and it can be a calculation using other variables
new_var_paste = stringr::str_glue("{hospital} on ({date_hospitalisation})") # new column = pasting together values from other columns
) %>%
select(case_id, hospital, date_hospitalisation, contains("new")) # show only new columns, for demonstration purposesReview the new columns. For demonstration purposes, only the new columns and the columns used to create them are shown:
TIP: A variation on mutate() is the function transmute(). This function adds a new column just like mutate(), but also drops/removes all other columns that you do not mention within its parentheses.
# HIDDEN FROM READER
# removes new demo columns created above
# linelist <- linelist %>%
# select(-contains("new_var"))Convert column class
Columns containing values that are dates, numbers, or logical values (TRUE/FALSE) will only behave as expected if they are correctly classified. There is a difference between “2” of class character and 2 of class numeric!
There are ways to set column class during the import commands, but this is often cumbersome. See the [R Basics] section on object classes to learn more about converting the class of objects and columns.
First, let’s run some checks on important columns to see if they are the correct class. We also saw this in the beginning when we ran skim().
Currently, the class of the age column is character. To perform quantitative analyses, we need these numbers to be recognized as numeric!
class(linelist$age)## [1] "character"The class of the date_onset column is also character! To perform analyses, these dates must be recognized as dates!
class(linelist$date_onset)## [1] "character"To resolve this, use the ability of mutate() to re-define a column with a transformation. We define the column as itself, but converted to a different class. Here is a basic example, converting or ensuring that the column age is class Numeric:
linelist <- linelist %>%
mutate(age = as.numeric(age))In a similar way, you can use as.character() and as.logical(). To convert to class Factor, you can use factor() from base R or as_factor() from forcats. Read more about this in the Factors page.
You must be careful when converting to class Date. Several methods are explained on the page Working with dates. Typically, the raw date values must all be in the same format for conversion to work correctly (e.g “MM/DD/YYYY”, or “DD MM YYYY”). After converting to class Date, check your data to confirm that each value was converted correctly.
Grouped data
If your data frame is already grouped (see page on Grouping data), mutate() may behave differently than if the data frame is not grouped. Any summarizing functions, like mean(), median(), max(), etc. will calculate by group, not by all the rows.
# age normalized to mean of ALL rows
linelist %>%
mutate(age_norm = age / mean(age, na.rm=T))
# age normalized to mean of hospital group
linelist %>%
group_by(hospital) %>%
mutate(age_norm = age / mean(age, na.rm=T))Read more about using mutate () on grouped dataframes in this tidyverse mutate documentation.
Transform multiple columns
Often to write concise code you want to apply the same transformation to multiple columns at once. A transformation can be applied to multiple columns at once using the across() function from the package dplyr (also contained within tidyverse package). across() can be used with any dplyr function, but is commonly used within select(), mutate(), filter(), or summarise(). See how it is applied to summarise() in the page on Descriptive tables.
Specify the columns to the argument .cols = and the function(s) to apply to .fns =. Any additional arguments to provide to the .fns function can be included after a comma, still within across().
across() column selection
Specify the columns to the argument .cols =. You can name them individually, or use “tidyselect” helper functions. Specify the function to .fns =. Note that using the function mode demonstrated below, the function is written without its parentheses ( ).
Here the transformation as.character() is applied to specific columns named within across().
linelist <- linelist %>%
mutate(across(.cols = c(temp, ht_cm, wt_kg), .fns = as.character))The “tidyselect” helper functions are available to assist you in specifying columns. They are detailed above in the section on Selecting and re-ordering columns, and they include: everything(), last_col(), where(), starts_with(), ends_with(), contains(), matches(), num_range() and any_of().
Here is an example of how one would change all columns to character class:
#to change all columns to character class
linelist <- linelist %>%
mutate(across(.cols = everything(), .fns = as.character))Convert to character all columns where the name contains the string “date” (note the placement of commas and parentheses):
#to change all columns to character class
linelist <- linelist %>%
mutate(across(.cols = contains("date"), .fns = as.character))Below, an example of mutating the columns that are currently class POSIXct (a raw datetime class that shows timestamps) - in other words, where the function is.POSIXct() evaluates to TRUE. Then we want to apply the function as.Date() to these columns to convert them to a normal class Date.
linelist <- linelist %>%
mutate(across(.cols = where(is.POSIXct), .fns = as.Date))- Note that within
across()we also use the functionwhere()asis.POSIXctis evaluating to either TRUE or FALSE.
- Note that
is.POSIXct()is from the package lubridate. Other similar “is” functions likeis.character(),is.numeric(), andis.logical()are from base R
across() functions
You can read the documentation with ?across for details on how to provide functions to across(). A few summary points: there are several ways to specify the function(s) to perform on a column and you can even define your own functions:
You can provide the function name alone (e.g.
meanoras.character)You can provide the function in purrr-style (e.g.
~ mean(.x, na.rm = TRUE)) (see this page)-
You can specify multiple functions by providing a list (e.g.
list(mean = mean, n_miss = ~ sum(is.na(.x))).- If you provide multiple functions, multiple transformed columns will be returned per input column, with unique names in the format
col_fn. You can adjust how the new columns are named with the.names =argument using glue syntax (see page on Characters and strings) where{.col}and{.fn}are shorthand for the input column and function.
- If you provide multiple functions, multiple transformed columns will be returned per input column, with unique names in the format
Here are a few online resources on using across(): creator Hadley Wickham’s thoughts/rationale
coalesce()
This dplyr function finds the first non-missing value at each position. It “fills-in” missing values with the first available value in an order you specify.
Here is an example outside the context of a data frame: Let us say you have two vectors, one containing the patient’s village of detection and another containing the patient’s village of residence. You can use coalesce to pick the first non-missing value for each index:
village_detection <- c("a", "b", NA, NA)
village_residence <- c("a", "c", "a", "d")
village <- coalesce(village_detection, village_residence)
village # print## [1] "a" "b" "a" "d"This works the same if you provide data frame columns: for each row, the function will assign the new column value with the first non-missing value in the columns you provided (in order provided).
linelist <- linelist %>%
mutate(village = coalesce(village_detection, village_residence))This is an example of a “row-wise” operation. For more complicated row-wise calculations, see the section below on Row-wise calculations.
Cumulative math
If you want a column to reflect the cumulative sum/mean/min/max etc as assessed down the rows of a dataframe to that point, use the following functions:
cumsum() returns the cumulative sum, as shown below:
## [1] 31## [1] 2 6 21 31This can be used in a dataframe when making a new column. For example, to calculate the cumulative number of cases per day in an outbreak, consider code like this:
cumulative_case_counts <- linelist %>% # begin with case linelist
count(date_onset) %>% # count of rows per day, as column 'n'
mutate(cumulative_cases = cumsum(n)) # new column, of the cumulative sum at each rowBelow are the first 10 rows:
head(cumulative_case_counts, 10)## date_onset n cumulative_cases
## 1 2012-04-15 1 1
## 2 2012-05-05 1 2
## 3 2012-05-08 1 3
## 4 2012-05-31 1 4
## 5 2012-06-02 1 5
## 6 2012-06-07 1 6
## 7 2012-06-14 1 7
## 8 2012-06-21 1 8
## 9 2012-06-24 1 9
## 10 2012-06-25 1 10See the page on Epidemic curves for how to plot cumulative incidence with the epicurve.
See also:cumsum(), cummean(), cummin(), cummax(), cumany(), cumall()
Using base R
To define a new column (or re-define a column) using base R, write the name of data frame, connected with $, to the new column (or the column to be modified). Use the assignment operator <- to define the new value(s). Remember that when using base R you must specify the data frame name before the column name every time (e.g. dataframe$column). Here is an example of creating the bmi column using base R:
linelist$bmi = linelist$wt_kg / (linelist$ht_cm / 100) ^ 2)Add to pipe chain
Below, a new column is added to the pipe chain and some classes are converted.
# CLEANING 'PIPE' CHAIN (starts with raw data and pipes it through cleaning steps)
##################################################################################
# begin cleaning pipe chain
###########################
linelist <- linelist_raw %>%
# standardize column name syntax
janitor::clean_names() %>%
# manually re-name columns
# NEW name # OLD name
rename(date_infection = infection_date,
date_hospitalisation = hosp_date,
date_outcome = date_of_outcome) %>%
# remove column
select(-c(row_num, merged_header, x28)) %>%
# de-duplicate
distinct() %>%
# ABOVE ARE UPSTREAM CLEANING STEPS ALREADY DISCUSSED
###################################################
# add new column
mutate(bmi = wt_kg / (ht_cm/100)^2) %>%
# convert class of columns
mutate(across(contains("date"), as.Date),
generation = as.numeric(generation),
age = as.numeric(age)) 8.8 Re-code values
Here are a few scenarios where you need to re-code (change) values:
- to edit one specific value (e.g. one date with an incorrect year or format)
- to reconcile values not spelled the same
- to create a new column of categorical values
- to create a new column of numeric categories (e.g. age categories)
Specific values
To change values manually you can use the recode() function within the mutate() function.
Imagine there is a nonsensical date in the data (e.g. “2014-14-15”): you could fix the date manually in the raw source data, or, you could write the change into the cleaning pipeline via mutate() and recode(). The latter is more transparent and reproducible to anyone else seeking to understand or repeat your analysis.
# fix incorrect values # old value # new value
linelist <- linelist %>%
mutate(date_onset = recode(date_onset, "2014-14-15" = "2014-04-15"))The mutate() line above can be read as: “mutate the column date_onset to equal the column date_onset re-coded so that OLD VALUE is changed to NEW VALUE”. Note that this pattern (OLD = NEW) for recode() is the opposite of most R patterns (new = old). The R development community is working on revising this.
Here is another example re-coding multiple values within one column.
In linelist the values in the column “hospital” must be cleaned. There are several different spellings and many missing values.
table(linelist$hospital, useNA = "always") # print table of all unique values, including missing ##
## Central Hopital Central Hospital Hospital A
## 11 457 290
## Hospital B Military Hopital Military Hospital
## 289 32 798
## Mitylira Hopital Mitylira Hospital Other
## 1 79 907
## Port Hopital Port Hospital St. Mark's Maternity Hospital (SMMH)
## 48 1756 417
## St. Marks Maternity Hopital (SMMH) <NA>
## 11 1512The recode() command below re-defines the column “hospital” as the current column “hospital”, but with the specified recode changes. Don’t forget commas after each!
linelist <- linelist %>%
mutate(hospital = recode(hospital,
# for reference: OLD = NEW
"Mitylira Hopital" = "Military Hospital",
"Mitylira Hospital" = "Military Hospital",
"Military Hopital" = "Military Hospital",
"Port Hopital" = "Port Hospital",
"Central Hopital" = "Central Hospital",
"other" = "Other",
"St. Marks Maternity Hopital (SMMH)" = "St. Mark's Maternity Hospital (SMMH)"
))Now we see the spellings in the hospital column have been corrected and consolidated:
table(linelist$hospital, useNA = "always")##
## Central Hospital Hospital A Hospital B
## 468 290 289
## Military Hospital Other Port Hospital
## 910 907 1804
## St. Mark's Maternity Hospital (SMMH) <NA>
## 428 1512TIP: The number of spaces before and after an equals sign does not matter. Make your code easier to read by aligning the = for all or most rows. Also, consider adding a hashed comment row to clarify for future readers which side is OLD and which side is NEW.
TIP: Sometimes a blank character value exists in a dataset (not recognized as R’s value for missing - NA). You can reference this value with two quotation marks with no space inbetween ("").
Simple logic
replace()
To re-code with simple logical criteria, you can use replace() within mutate(). replace() is a function from base R. Use a logic condition to specify the rows to change . The general syntax is:
mutate(col_to_change = replace(col_to_change, criteria for rows, new value)).
One common situation to use replace() is changing just one value in one row, using an unique row identifier. Below, the gender is changed to “Female” in the row where the column case_id is “2195”.
# Example: change gender of one specific observation to "Female"
linelist <- linelist %>%
mutate(gender = replace(gender, case_id == "2195", "Female"))The equivalent command using base R syntax and indexing brackets [ ] is below. It reads as “Change the value of the dataframe linelist‘s column gender (for the rows where linelist’s column case_id has the value ’2195’) to ‘Female’”.
linelist$gender[linelist$case_id == "2195"] <- "Female"
ifelse() and if_else()
Another tool for simple logic is ifelse() and its partner if_else(). However, in most cases for re-coding it is more clear to use case_when() (detailed below). These “if else” commands are simplified versions of an if and else programming statement. The general syntax is:ifelse(condition, value to return if condition evaluates to TRUE, value to return if condition evaluates to FALSE)
Below, the column source_known is defined. Its value in a given row is set to “known” if the row’s value in column source is not missing. If the value in source is missing, then the value in source_known is set to “unknown”.
if_else() is a special version from dplyr that handles dates. Note that if the ‘true’ value is a date, the ‘false’ value must also qualify a date, hence using the special value NA_real_ instead of just NA.
# Create a date of death column, which is NA if patient has not died.
linelist <- linelist %>%
mutate(date_death = if_else(outcome == "Death", date_outcome, NA_real_))Avoid stringing together many ifelse commands… use case_when() instead! case_when() is much easier to read and you’ll make fewer errors.
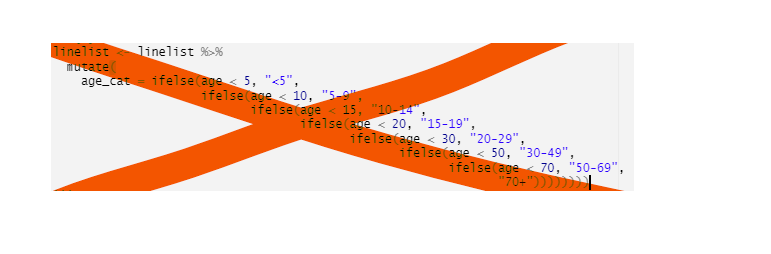
Outside of the context of a data frame, if you want to have an object used in your code switch its value, consider using switch() from base R.
Complex logic
Use dplyr’s case_when() if you are re-coding into many new groups, or if you need to use complex logic statements to re-code values. This function evaluates every row in the data frame, assess whether the rows meets specified criteria, and assigns the correct new value.
case_when() commands consist of statements that have a Right-Hand Side (RHS) and a Left-Hand Side (LHS) separated by a “tilde” ~. The logic criteria are in the left side and the pursuant values are in the right side of each statement. Statements are separated by commas.
For example, here we utilize the columns age and age_unit to create a column age_years:
linelist <- linelist %>%
mutate(age_years = case_when(
age_unit == "years" ~ age, # if age is given in years
age_unit == "months" ~ age/12, # if age is given in months
is.na(age_unit) ~ age, # if age unit is missing, assume years
TRUE ~ NA_real_)) # any other circumstance, assign missingAs each row in the data is evaluated, the criteria are applied/evaluated in the order the case_when() statements are written - from top-to-bottom. If the top criteria evaluates to TRUE for a given row, the RHS value is assigned, and the remaining criteria are not even tested for that row. Thus, it is best to write the most specific criteria first, and the most general last.
Along those lines, in your final statement, place TRUE on the left-side, which will capture any row that did not meet any of the previous criteria. The right-side of this statement could be assigned a value like “check me!” or missing.
DANGER: Vvalues on the right-side must all be the same class - either numeric, character, date, logical, etc. To assign missing (NA), you may need to use special variations of NA such as NA_character_, NA_real_ (for numeric or POSIX), and as.Date(NA). Read more in Working with dates.
Missing values
Below are special functions for handling missing values in the context of data cleaning.
See the page on Missing data for more detailed tips on identifying and handling missing values. For example, the is.na() function which logically tests for missingness.
replace_na()
To change missing values (NA) to a specific value, such as “Missing”, use the dplyr function replace_na() within mutate(). Note that this is used in the same manner as recode above - the name of the variable must be repeated within replace_na().
linelist <- linelist %>%
mutate(hospital = replace_na(hospital, "Missing"))fct_explicit_na()
This is a function from the forcats package. The forcats package handles columns of class Factor. Factors are R’s way to handle ordered values such as c("First", "Second", "Third") or to set the order that values (e.g. hospitals) appear in tables and plots. See the page on Factors.
If your data are class Factor and you try to convert NA to “Missing” by using replace_na(), you will get this error: invalid factor level, NA generated. You have tried to add “Missing” as a value, when it was not defined as a possible level of the factor, and it was rejected.
The easiest way to solve this is to use the forcats function fct_explicit_na() which converts a column to class factor, and converts NA values to the character “(Missing)”.
linelist %>%
mutate(hospital = fct_explicit_na(hospital))A slower alternative would be to add the factor level using fct_expand() and then convert the missing values.
na_if()
To convert a specific value to NA, use dplyr’s na_if(). The command below performs the opposite operation of replace_na(). In the example below, any values of “Missing” in the column hospital are converted to NA.
linelist <- linelist %>%
mutate(hospital = na_if(hospital, "Missing"))Note: na_if() cannot be used for logic criteria (e.g. “all values > 99”) - use replace() or case_when() for this:
Cleaning dictionary
Use the R package linelist and it’s function clean_variable_spelling() to clean a data frame with a cleaning dictionary. linelist is a package developed by RECON - the R Epidemics Consortium.
-
Create a cleaning dictionary with 3 columns:
- A “from” column (the incorrect value)
- A “to” column (the correct value)
- A column specifying the column for the changes to be applied (or “.global” to apply to all columns)
- A “from” column (the incorrect value)
Note: .global dictionary entries will be overridden by column-specific dictionary entries.
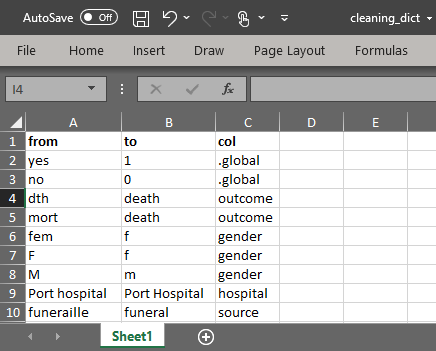
- Import the dictionary file into R. This example can be downloaded via instructions on the [Download handbook and data] page.
cleaning_dict <- import("cleaning_dict.csv")- Pass the raw linelist to
clean_variable_spelling(), specifying towordlists =the cleaning dictionary data frame. Thespelling_vars =argument can be used to specify which column in the dictionary refers to the columns (3rd by default), or can be set toNULLto have the dictionary apply to all character and factor columns. Note this function can take a long time to run.
linelist <- linelist %>%
linelist::clean_variable_spelling(
wordlists = cleaning_dict,
spelling_vars = "col", # dict column containing column names, defaults to 3rd column in dict
)Now scroll to the right to see how values have changed - particularly gender (lowercase to uppercase), and all the symptoms columns have been transformed from yes/no to 1/0.
Note that your column names in the cleaning dictionary must correspond to the names at this point in your cleaning script. See this online reference for the linelist package for more details.
Add to pipe chain
Below, some new columns and column transformations are added to the pipe chain.
# CLEANING 'PIPE' CHAIN (starts with raw data and pipes it through cleaning steps)
##################################################################################
# begin cleaning pipe chain
###########################
linelist <- linelist_raw %>%
# standardize column name syntax
janitor::clean_names() %>%
# manually re-name columns
# NEW name # OLD name
rename(date_infection = infection_date,
date_hospitalisation = hosp_date,
date_outcome = date_of_outcome) %>%
# remove column
select(-c(row_num, merged_header, x28)) %>%
# de-duplicate
distinct() %>%
# add column
mutate(bmi = wt_kg / (ht_cm/100)^2) %>%
# convert class of columns
mutate(across(contains("date"), as.Date),
generation = as.numeric(generation),
age = as.numeric(age)) %>%
# add column: delay to hospitalisation
mutate(days_onset_hosp = as.numeric(date_hospitalisation - date_onset)) %>%
# ABOVE ARE UPSTREAM CLEANING STEPS ALREADY DISCUSSED
###################################################
# clean values of hospital column
mutate(hospital = recode(hospital,
# OLD = NEW
"Mitylira Hopital" = "Military Hospital",
"Mitylira Hospital" = "Military Hospital",
"Military Hopital" = "Military Hospital",
"Port Hopital" = "Port Hospital",
"Central Hopital" = "Central Hospital",
"other" = "Other",
"St. Marks Maternity Hopital (SMMH)" = "St. Mark's Maternity Hospital (SMMH)"
)) %>%
mutate(hospital = replace_na(hospital, "Missing")) %>%
# create age_years column (from age and age_unit)
mutate(age_years = case_when(
age_unit == "years" ~ age,
age_unit == "months" ~ age/12,
is.na(age_unit) ~ age,
TRUE ~ NA_real_))8.9 Numeric categories
Here we describe some special approaches for creating categories from numerical columns. Common examples include age categories, groups of lab values, etc. Here we will discuss:
-
age_categories(), from the epikit package
-
cut(), from base R
-
case_when()
- quantile breaks with
quantile()andntile()
Review distribution
For this example we will create an age_cat column using the age_years column.
#check the class of the linelist variable age
class(linelist$age_years)## [1] "numeric"First, examine the distribution of your data, to make appropriate cut-points. See the page on ggplot basics.
# examine the distribution
hist(linelist$age_years)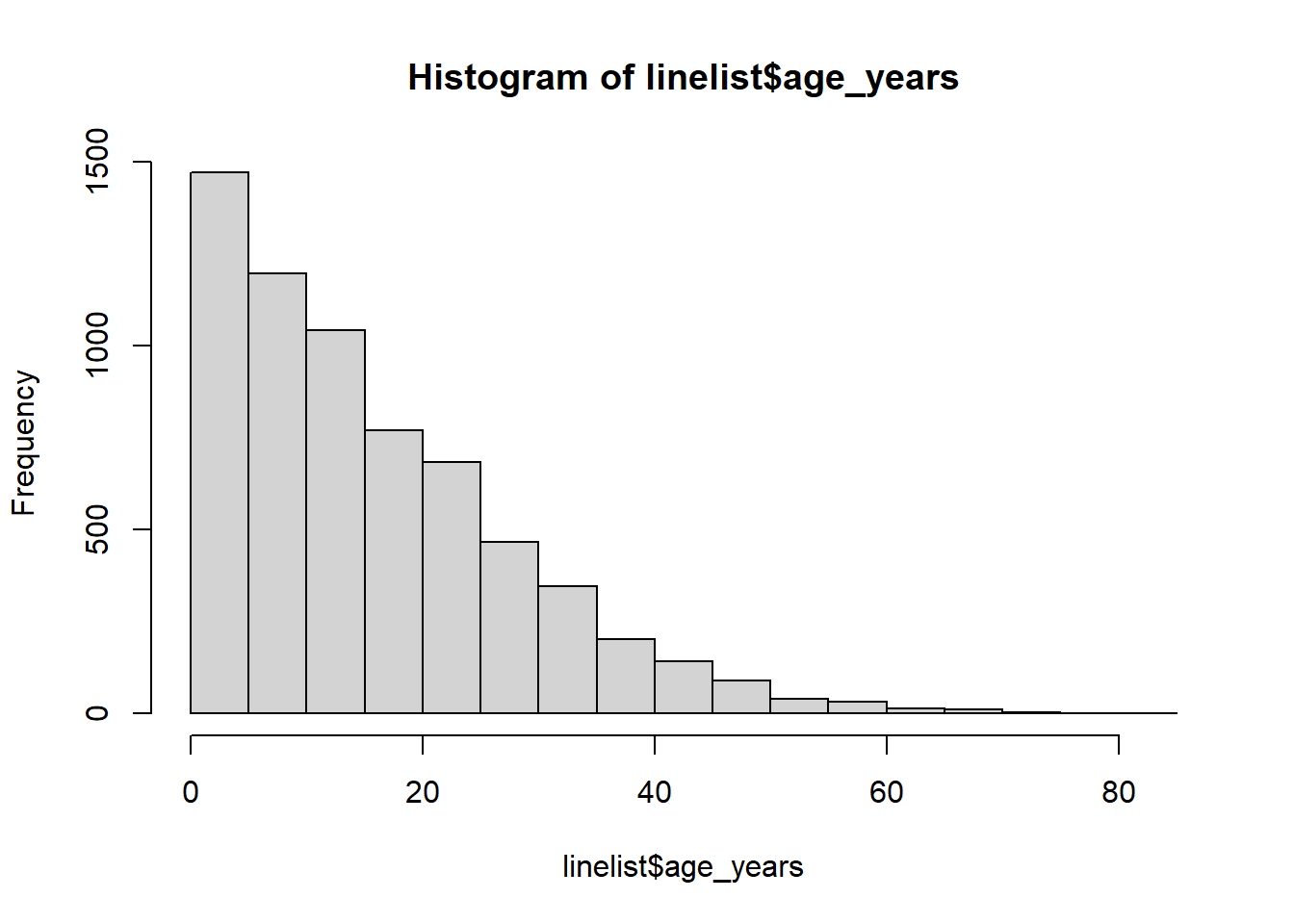
summary(linelist$age_years, na.rm=T)## Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
## 0.00 6.00 13.00 16.04 23.00 84.00 107CAUTION: Sometimes, numeric variables will import as class “character”. This occurs if there are non-numeric characters in some of the values, for example an entry of “2 months” for age, or (depending on your R locale settings) if a comma is used in the decimals place (e.g. “4,5” to mean four and one half years)..
age_categories()
With the epikit package, you can use the age_categories() function to easily categorize and label numeric columns (note: this function can be applied to non-age numeric variables too). As a bonum, the output column is automatically an ordered factor.
Here are the required inputs:
- A numeric vector (column)
- The
breakers =argument - provide a numeric vector of break points for the new groups
First, the simplest example:
# Simple example
################
pacman::p_load(epikit) # load package
linelist <- linelist %>%
mutate(
age_cat = age_categories( # create new column
age_years, # numeric column to make groups from
breakers = c(0, 5, 10, 15, 20, # break points
30, 40, 50, 60, 70)))
# show table
table(linelist$age_cat, useNA = "always")##
## 0-4 5-9 10-14 15-19 20-29 30-39 40-49 50-59 60-69 70+ <NA>
## 1227 1223 1048 827 1216 597 251 78 27 7 107The break values you specify are by default the lower bounds - that is, they are included in the “higher” group / the groups are “open” on the lower/left side. As shown below, you can add 1 to each break value to achieve groups that are open at the top/right.
# Include upper ends for the same categories
############################################
linelist <- linelist %>%
mutate(
age_cat = age_categories(
age_years,
breakers = c(0, 6, 11, 16, 21, 31, 41, 51, 61, 71)))
# show table
table(linelist$age_cat, useNA = "always")##
## 0-5 6-10 11-15 16-20 21-30 31-40 41-50 51-60 61-70 71+ <NA>
## 1469 1195 1040 770 1149 547 231 70 24 6 107You can adjust how the labels are displayed with separator =. The default is “-”
You can adjust how the top numbers are handled, with the ceiling = arguemnt. To set an upper cut-off set ceiling = TRUE. In this use, the highest break value provided is a “ceiling” and a category “XX+” is not created. Any values above highest break value (or to upper =, if defined) are categorized as NA. Below is an example with ceiling = TRUE, so that there is no category of XX+ and values above 70 (the highest break value) are assigned as NA.
# With ceiling set to TRUE
##########################
linelist <- linelist %>%
mutate(
age_cat = age_categories(
age_years,
breakers = c(0, 5, 10, 15, 20, 30, 40, 50, 60, 70),
ceiling = TRUE)) # 70 is ceiling, all above become NA
# show table
table(linelist$age_cat, useNA = "always")##
## 0-4 5-9 10-14 15-19 20-29 30-39 40-49 50-59 60-70 <NA>
## 1227 1223 1048 827 1216 597 251 78 28 113Alternatively, instead of breakers =, you can provide all of lower =, upper =, and by =:
-
lower =The lowest number you want considered - default is 0
-
upper =The highest number you want considered
-
by =The number of years between groups
linelist <- linelist %>%
mutate(
age_cat = age_categories(
age_years,
lower = 0,
upper = 100,
by = 10))
# show table
table(linelist$age_cat, useNA = "always")##
## 0-9 10-19 20-29 30-39 40-49 50-59 60-69 70-79 80-89 90-99 100+ <NA>
## 2450 1875 1216 597 251 78 27 6 1 0 0 107See the function’s Help page for more details (enter ?age_categories in the R console).
cut()
cut() is a base R alternative to age_categories(), but I think you will see why age_categories() was developed to simplify this process. Some notable differences from age_categories() are:
- You do not need to install/load another package
- You can specify whether groups are open/closed on the right/left
- You must provide accurate labels yourself
- If you want 0 included in the lowest group you must specify this
The basic syntax within cut() is to first provide the numeric column to be cut (age_years), and then the breaks argument, which is a numeric vector c() of break points. Using cut(), the resulting column is an ordered factor.
By default, the categorization occurs so that the right/upper side is “open” and inclusive (and the left/lower side is “closed” or exclusive). This is the opposite behavior from the age_categories() function. The default labels use the notation “(A, B]”, which means A is not included but B is. Reverse this behavior by providing the right = TRUE argument.
Thus, by default, “0” values are excluded from the lowest group, and categorized as NA! “0” values could be infants coded as age 0 so be careful! To change this, add the argument include.lowest = TRUE so that any “0” values will be included in the lowest group. The automatically-generated label for the lowest category will then be “[A],B]”. Note that if you include the include.lowest = TRUE argument and right = TRUE, the extreme inclusion will now apply to the highest break point value and category, not the lowest.
You can provide a vector of customized labels using the labels = argument. As these are manually written, be very careful to ensure they are accurate! Check your work using cross-tabulation, as described below.
An example of cut() applied to age_years to make the new variable age_cat is below:
# Create new variable, by cutting the numeric age variable
# lower break is excluded but upper break is included in each category
linelist <- linelist %>%
mutate(
age_cat = cut(
age_years,
breaks = c(0, 5, 10, 15, 20,
30, 50, 70, 100),
include.lowest = TRUE # include 0 in lowest group
))
# tabulate the number of observations per group
table(linelist$age_cat, useNA = "always")##
## [0,5] (5,10] (10,15] (15,20] (20,30] (30,50] (50,70] (70,100] <NA>
## 1469 1195 1040 770 1149 778 94 6 107Check your work!!! Verify that each age value was assigned to the correct category by cross-tabulating the numeric and category columns. Examine assignment of boundary values (e.g. 15, if neighboring categories are 10-15 and 16-20).
# Cross tabulation of the numeric and category columns.
table("Numeric Values" = linelist$age_years, # names specified in table for clarity.
"Categories" = linelist$age_cat,
useNA = "always") # don't forget to examine NA values## Categories
## Numeric Values [0,5] (5,10] (10,15] (15,20] (20,30] (30,50] (50,70] (70,100] <NA>
## 0 136 0 0 0 0 0 0 0 0
## 0.0833333333333333 1 0 0 0 0 0 0 0 0
## 0.25 2 0 0 0 0 0 0 0 0
## 0.333333333333333 6 0 0 0 0 0 0 0 0
## 0.416666666666667 1 0 0 0 0 0 0 0 0
## 0.5 6 0 0 0 0 0 0 0 0
## 0.583333333333333 3 0 0 0 0 0 0 0 0
## 0.666666666666667 3 0 0 0 0 0 0 0 0
## 0.75 3 0 0 0 0 0 0 0 0
## 0.833333333333333 1 0 0 0 0 0 0 0 0
## 0.916666666666667 1 0 0 0 0 0 0 0 0
## 1 275 0 0 0 0 0 0 0 0
## 1.5 2 0 0 0 0 0 0 0 0
## 2 308 0 0 0 0 0 0 0 0
## 3 246 0 0 0 0 0 0 0 0
## 4 233 0 0 0 0 0 0 0 0
## 5 242 0 0 0 0 0 0 0 0
## 6 0 241 0 0 0 0 0 0 0
## 7 0 256 0 0 0 0 0 0 0
## 8 0 239 0 0 0 0 0 0 0
## 9 0 245 0 0 0 0 0 0 0
## 10 0 214 0 0 0 0 0 0 0
## 11 0 0 220 0 0 0 0 0 0
## 12 0 0 224 0 0 0 0 0 0
## 13 0 0 191 0 0 0 0 0 0
## 14 0 0 199 0 0 0 0 0 0
## 15 0 0 206 0 0 0 0 0 0
## 16 0 0 0 186 0 0 0 0 0
## 17 0 0 0 164 0 0 0 0 0
## 18 0 0 0 141 0 0 0 0 0
## 19 0 0 0 130 0 0 0 0 0
## 20 0 0 0 149 0 0 0 0 0
## 21 0 0 0 0 158 0 0 0 0
## 22 0 0 0 0 149 0 0 0 0
## 23 0 0 0 0 125 0 0 0 0
## 24 0 0 0 0 144 0 0 0 0
## 25 0 0 0 0 107 0 0 0 0
## 26 0 0 0 0 100 0 0 0 0
## 27 0 0 0 0 117 0 0 0 0
## 28 0 0 0 0 85 0 0 0 0
## 29 0 0 0 0 82 0 0 0 0
## 30 0 0 0 0 82 0 0 0 0
## 31 0 0 0 0 0 68 0 0 0
## 32 0 0 0 0 0 84 0 0 0
## 33 0 0 0 0 0 78 0 0 0
## 34 0 0 0 0 0 58 0 0 0
## 35 0 0 0 0 0 58 0 0 0
## 36 0 0 0 0 0 33 0 0 0
## 37 0 0 0 0 0 46 0 0 0
## 38 0 0 0 0 0 45 0 0 0
## 39 0 0 0 0 0 45 0 0 0
## 40 0 0 0 0 0 32 0 0 0
## 41 0 0 0 0 0 34 0 0 0
## 42 0 0 0 0 0 26 0 0 0
## 43 0 0 0 0 0 31 0 0 0
## 44 0 0 0 0 0 24 0 0 0
## 45 0 0 0 0 0 27 0 0 0
## 46 0 0 0 0 0 25 0 0 0
## 47 0 0 0 0 0 16 0 0 0
## 48 0 0 0 0 0 21 0 0 0
## 49 0 0 0 0 0 15 0 0 0
## 50 0 0 0 0 0 12 0 0 0
## 51 0 0 0 0 0 0 13 0 0
## 52 0 0 0 0 0 0 7 0 0
## 53 0 0 0 0 0 0 4 0 0
## 54 0 0 0 0 0 0 6 0 0
## 55 0 0 0 0 0 0 9 0 0
## 56 0 0 0 0 0 0 7 0 0
## 57 0 0 0 0 0 0 9 0 0
## 58 0 0 0 0 0 0 6 0 0
## 59 0 0 0 0 0 0 5 0 0
## 60 0 0 0 0 0 0 4 0 0
## 61 0 0 0 0 0 0 2 0 0
## 62 0 0 0 0 0 0 1 0 0
## 63 0 0 0 0 0 0 5 0 0
## 64 0 0 0 0 0 0 1 0 0
## 65 0 0 0 0 0 0 5 0 0
## 66 0 0 0 0 0 0 3 0 0
## 67 0 0 0 0 0 0 2 0 0
## 68 0 0 0 0 0 0 1 0 0
## 69 0 0 0 0 0 0 3 0 0
## 70 0 0 0 0 0 0 1 0 0
## 72 0 0 0 0 0 0 0 1 0
## 73 0 0 0 0 0 0 0 3 0
## 76 0 0 0 0 0 0 0 1 0
## 84 0 0 0 0 0 0 0 1 0
## <NA> 0 0 0 0 0 0 0 0 107Re-labeling NA values
You may want to assign NA values a label such as “Missing”. Because the new column is class Factor (restricted values), you cannot simply mutate it with replace_na(), as this value will be rejected. Instead, use fct_explicit_na() from forcats as explained in the Factors page.
linelist <- linelist %>%
# cut() creates age_cat, automatically of class Factor
mutate(age_cat = cut(
age_years,
breaks = c(0, 5, 10, 15, 20, 30, 50, 70, 100),
right = FALSE,
include.lowest = TRUE,
labels = c("0-4", "5-9", "10-14", "15-19", "20-29", "30-49", "50-69", "70-100")),
# make missing values explicit
age_cat = fct_explicit_na(
age_cat,
na_level = "Missing age") # you can specify the label
)
# table to view counts
table(linelist$age_cat, useNA = "always")##
## 0-4 5-9 10-14 15-19 20-29 30-49 50-69 70-100 Missing age <NA>
## 1227 1223 1048 827 1216 848 105 7 107 0Quickly make breaks and labels
For a fast way to make breaks and label vectors, use something like below. See the [R basics] page for references on seq() and rep().
# Make break points from 0 to 90 by 5
age_seq = seq(from = 0, to = 90, by = 5)
age_seq
# Make labels for the above categories, assuming default cut() settings
age_labels = paste0(age_seq + 1, "-", age_seq + 5)
age_labels
# check that both vectors are the same length
length(age_seq) == length(age_labels)Read more about cut() in its Help page by entering ?cut in the R console.
Quantile breaks
In common understanding, “quantiles” or “percentiles” typically refer to a value below which a proportion of values fall. For example, the 95th percentile of ages in linelist would be the age below which 95% of the age fall.
However in common speech, “quartiles” and “deciles” can also refer to the groups of data as equally divided into 4, or 10 groups (note there will be one more break point than group).
To get quantile break points, you can use quantile() from the stats package from base R. You provide a numeric vector (e.g. a column in a dataset) and vector of numeric probability values ranging from 0 to 1.0. The break points are returned as a numeric vector. Explore the details of the statistical methodologies by entering ?quantile.
- If your input numeric vector has any missing values it is best to set
na.rm = TRUE
- Set
names = FALSEto get an un-named numeric vector
quantile(linelist$age_years, # specify numeric vector to work on
probs = c(0, .25, .50, .75, .90, .95), # specify the percentiles you want
na.rm = TRUE) # ignore missing values ## 0% 25% 50% 75% 90% 95%
## 0 6 13 23 33 41You can use the results of quantile() as break points in age_categories() or cut(). Below we create a new column deciles using cut() where the breaks are defined using quantiles() on age_years. Below, we display the results using tabyl() from janitor so you can see the percentages (see the Descriptive tables page). Note how they are not exactly 10% in each group.
linelist %>% # begin with linelist
mutate(deciles = cut(age_years, # create new column decile as cut() on column age_years
breaks = quantile( # define cut breaks using quantile()
age_years, # operate on age_years
probs = seq(0, 1, by = 0.1), # 0.0 to 1.0 by 0.1
na.rm = TRUE), # ignore missing values
include.lowest = TRUE)) %>% # for cut() include age 0
janitor::tabyl(deciles) # pipe to table to display## deciles n percent valid_percent
## [0,2] 748 0.11319613 0.11505922
## (2,5] 721 0.10911017 0.11090601
## (5,7] 497 0.07521186 0.07644978
## (7,10] 698 0.10562954 0.10736810
## (10,13] 635 0.09609564 0.09767728
## (13,17] 755 0.11425545 0.11613598
## (17,21] 578 0.08746973 0.08890940
## (21,26] 625 0.09458232 0.09613906
## (26,33] 596 0.09019370 0.09167820
## (33,84] 648 0.09806295 0.09967697
## <NA> 107 0.01619249 NAEvenly-sized groups
Another tool to make numeric groups is the the dplyr function ntile(), which attempts to break your data into n evenly-sized groups - but be aware that unlike with quantile() the same value could appear in more than one group. Provide the numeric vector and then the number of groups. The values in the new column created is just group “numbers” (e.g. 1 to 10), not the range of values themselves as when using cut().
# make groups with ntile()
ntile_data <- linelist %>%
mutate(even_groups = ntile(age_years, 10))
# make table of counts and proportions by group
ntile_table <- ntile_data %>%
janitor::tabyl(even_groups)
# attach min/max values to demonstrate ranges
ntile_ranges <- ntile_data %>%
group_by(even_groups) %>%
summarise(
min = min(age_years, na.rm=T),
max = max(age_years, na.rm=T)
)## Warning in min(age_years, na.rm = T): no non-missing arguments to min; returning Inf## Warning in max(age_years, na.rm = T): no non-missing arguments to max; returning -Inf
# combine and print - note that values are present in multiple groups
left_join(ntile_table, ntile_ranges, by = "even_groups")## even_groups n percent valid_percent min max
## 1 651 0.09851695 0.10013844 0 2
## 2 650 0.09836562 0.09998462 2 5
## 3 650 0.09836562 0.09998462 5 7
## 4 650 0.09836562 0.09998462 7 10
## 5 650 0.09836562 0.09998462 10 13
## 6 650 0.09836562 0.09998462 13 17
## 7 650 0.09836562 0.09998462 17 21
## 8 650 0.09836562 0.09998462 21 26
## 9 650 0.09836562 0.09998462 26 33
## 10 650 0.09836562 0.09998462 33 84
## NA 107 0.01619249 NA Inf -Inf
case_when()
It is possible to use the dplyr function case_when() to create categories from a numeric column, but it is easier to use age_categories() from epikit or cut() because these will create an ordered factor automatically.
If using case_when(), please review the proper use as described earlier in the Re-code values section of this page. Also be aware that all right-hand side values must be of the same class. Thus, if you want NA on the right-side you should either write “Missing” or use the special NA value NA_character_.
Add to pipe chain
Below, code to create two categorical age columns is added to the cleaning pipe chain:
# CLEANING 'PIPE' CHAIN (starts with raw data and pipes it through cleaning steps)
##################################################################################
# begin cleaning pipe chain
###########################
linelist <- linelist_raw %>%
# standardize column name syntax
janitor::clean_names() %>%
# manually re-name columns
# NEW name # OLD name
rename(date_infection = infection_date,
date_hospitalisation = hosp_date,
date_outcome = date_of_outcome) %>%
# remove column
select(-c(row_num, merged_header, x28)) %>%
# de-duplicate
distinct() %>%
# add column
mutate(bmi = wt_kg / (ht_cm/100)^2) %>%
# convert class of columns
mutate(across(contains("date"), as.Date),
generation = as.numeric(generation),
age = as.numeric(age)) %>%
# add column: delay to hospitalisation
mutate(days_onset_hosp = as.numeric(date_hospitalisation - date_onset)) %>%
# clean values of hospital column
mutate(hospital = recode(hospital,
# OLD = NEW
"Mitylira Hopital" = "Military Hospital",
"Mitylira Hospital" = "Military Hospital",
"Military Hopital" = "Military Hospital",
"Port Hopital" = "Port Hospital",
"Central Hopital" = "Central Hospital",
"other" = "Other",
"St. Marks Maternity Hopital (SMMH)" = "St. Mark's Maternity Hospital (SMMH)"
)) %>%
mutate(hospital = replace_na(hospital, "Missing")) %>%
# create age_years column (from age and age_unit)
mutate(age_years = case_when(
age_unit == "years" ~ age,
age_unit == "months" ~ age/12,
is.na(age_unit) ~ age,
TRUE ~ NA_real_)) %>%
# ABOVE ARE UPSTREAM CLEANING STEPS ALREADY DISCUSSED
###################################################
mutate(
# age categories: custom
age_cat = epikit::age_categories(age_years, breakers = c(0, 5, 10, 15, 20, 30, 50, 70)),
# age categories: 0 to 85 by 5s
age_cat5 = epikit::age_categories(age_years, breakers = seq(0, 85, 5)))8.10 Add rows
One-by-one
Adding rows one-by-one manually is tedious but can be done with add_row() from dplyr. Remember that each column must contain values of only one class (either character, numeric, logical, etc.). So adding a row requires nuance to maintain this.
linelist <- linelist %>%
add_row(row_num = 666,
case_id = "abc",
generation = 4,
`infection date` = as.Date("2020-10-10"),
.before = 2)Use .before and .after. to specify the placement of the row you want to add. .before = 3 will put the new row before the current 3rd row. The default behavior is to add the row to the end. Columns not specified will be left empty (NA).
The new row number may look strange (“…23”) but the row numbers in the pre-existing rows have changed. So if using the command twice, examine/test the insertion carefully.
If a class you provide is off you will see an error like this:
Error: Can't combine ..1$infection date <date> and ..2$infection date <character>.(when inserting a row with a date value, remember to wrap the date in the function as.Date() like as.Date("2020-10-10")).
Bind rows
To combine datasets together by binding the rows of one dataframe to the bottom of another data frame, you can use bind_rows() from dplyr. This is explained in more detail in the page Joining data.
8.11 Filter rows
A typical cleaning step after you have cleaned the columns and re-coded values is to filter the data frame for specific rows using the dplyr verb filter().
Within filter(), specify the logic that must be TRUE for a row in the dataset to be kept. Below we show how to filter rows based on simple and complex logical conditions.
Simple filter
This simple example re-defines the dataframe linelist as itself, having filtered the rows to meet a logical condition. Only the rows where the logical statement within the parentheses evaluates to TRUE are kept.
In this example, the logical statement is gender == "f", which is asking whether the value in the column gender is equal to “f” (case sensitive).
Before the filter is applied, the number of rows in linelist is nrow(linelist).
linelist <- linelist %>%
filter(gender == "f") # keep only rows where gender is equal to "f"After the filter is applied, the number of rows in linelist is linelist %>% filter(gender == "f") %>% nrow().
Filter out missing values
It is fairly common to want to filter out rows that have missing values. Resist the urge to write filter(!is.na(column) & !is.na(column)) and instead use the tidyr function that is custom-built for this purpose: drop_na(). If run with empty parentheses, it removes rows with any missing values. Alternatively, you can provide names of specific columns to be evaluated for missingness, or use the “tidyselect” helper functions described above.
linelist %>%
drop_na(case_id, age_years) # drop rows with missing values for case_id or age_yearsSee the page on Missing data for many techniques to analyse and manage missingness in your data.
Filter by row number
In a data frame or tibble, each row will usually have a “row number” that (when seen in R Viewer) appears to the left of the first column. It is not itself a true column in the data, but it can be used in a filter() statement.
To filter based on “row number”, you can use the dplyr function row_number() with open parentheses as part of a logical filtering statement. Often you will use the %in% operator and a range of numbers as part of that logical statement, as shown below. To see the first N rows, you can also use the special dplyr function head().
# View first 100 rows
linelist %>% head(100) # or use tail() to see the n last rows
# Show row 5 only
linelist %>% filter(row_number() == 5)
# View rows 2 through 20, and three specific columns
linelist %>% filter(row_number() %in% 2:20) %>% select(date_onset, outcome, age)You can also convert the row numbers to a true column by piping your data frame to the tibble function rownames_to_column() (do not put anything in the parentheses).
Complex filter
More complex logical statements can be constructed using parentheses ( ), OR |, negate !, %in%, and AND & operators. An example is below:
Note: You can use the ! operator in front of a logical criteria to negate it. For example, !is.na(column) evaluates to true if the column value is not missing. Likewise !column %in% c("a", "b", "c") evaluates to true if the column value is not in the vector.
Examine the data
Below is a simple one-line command to create a histogram of onset dates. See that a second smaller outbreak from 2012-2013 is also included in this raw dataset. For our analyses, we want to remove entries from this earlier outbreak.
hist(linelist$date_onset, breaks = 50)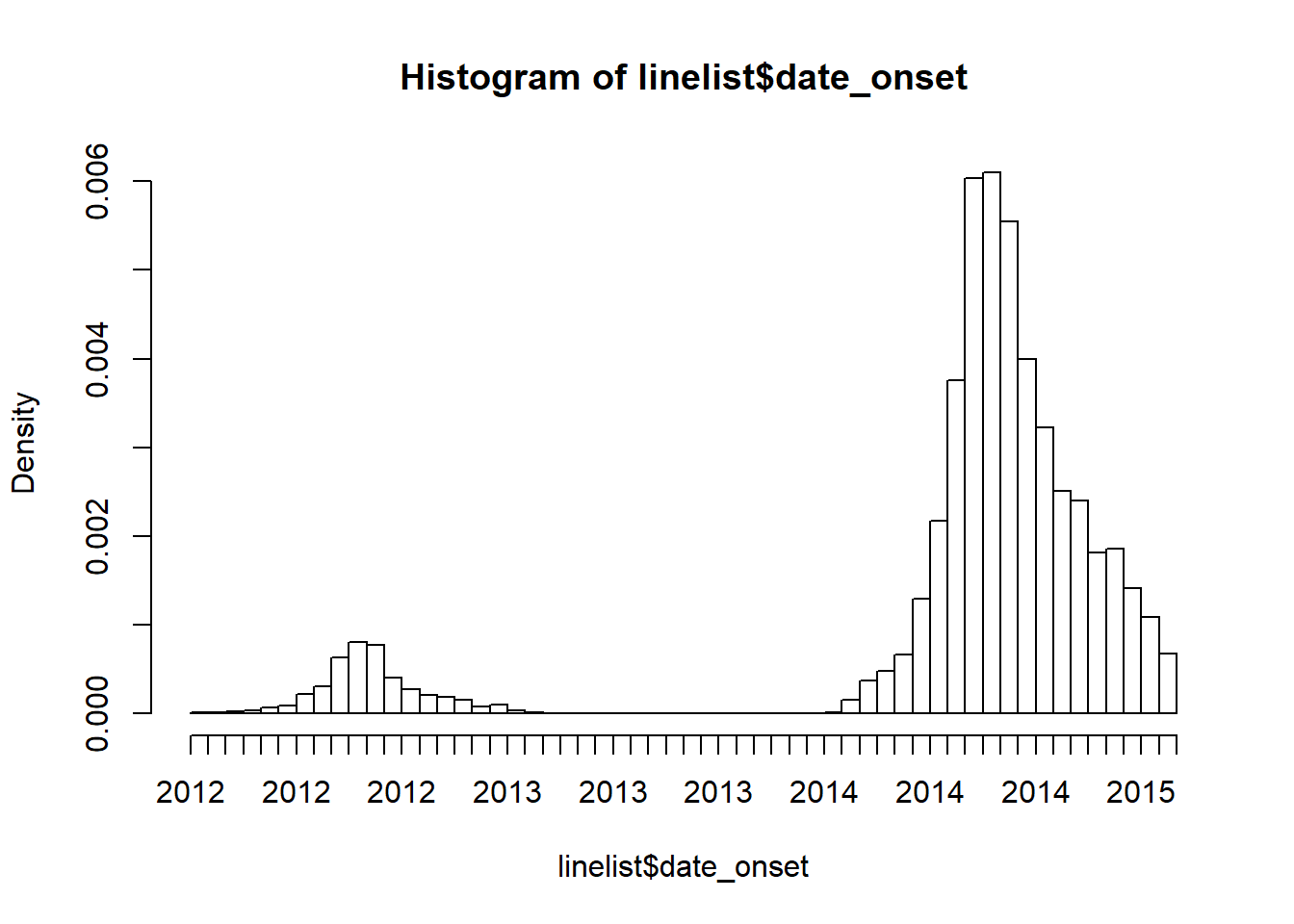
How filters handle missing numeric and date values
Can we just filter by date_onset to rows after June 2013? Caution! Applying the code filter(date_onset > as.Date("2013-06-01"))) would remove any rows in the later epidemic with a missing date of onset!
DANGER: Filtering to greater than (>) or less than (<) a date or number can remove any rows with missing values (NA)! This is because NA is treated as infinitely large and small.
(See the page on Working with dates for more information on working with dates and the package lubridate)
Design the filter
Examine a cross-tabulation to make sure we exclude only the correct rows:
table(Hospital = linelist$hospital, # hospital name
YearOnset = lubridate::year(linelist$date_onset), # year of date_onset
useNA = "always") # show missing values## YearOnset
## Hospital 2012 2013 2014 2015 <NA>
## Central Hospital 0 0 351 99 18
## Hospital A 229 46 0 0 15
## Hospital B 227 47 0 0 15
## Military Hospital 0 0 676 200 34
## Missing 0 0 1117 318 77
## Other 0 0 684 177 46
## Port Hospital 9 1 1372 347 75
## St. Mark's Maternity Hospital (SMMH) 0 0 322 93 13
## <NA> 0 0 0 0 0What other criteria can we filter on to remove the first outbreak (in 2012 & 2013) from the dataset? We see that:
- The first epidemic in 2012 & 2013 occurred at Hospital A, Hospital B, and that there were also 10 cases at Port Hospital.
- Hospitals A & B did not have cases in the second epidemic, but Port Hospital did.
We want to exclude:
-
The
nrow(linelist %>% filter(hospital %in% c("Hospital A", "Hospital B") | date_onset < as.Date("2013-06-01")))rows with onset in 2012 and 2013 at either hospital A, B, or Port:- Exclude
nrow(linelist %>% filter(date_onset < as.Date("2013-06-01")))rows with onset in 2012 and 2013 - Exclude
nrow(linelist %>% filter(hospital %in% c('Hospital A', 'Hospital B') & is.na(date_onset)))rows from Hospitals A & B with missing onset dates
- Do not exclude
nrow(linelist %>% filter(!hospital %in% c('Hospital A', 'Hospital B') & is.na(date_onset)))other rows with missing onset dates.
- Exclude
We start with a linelist of nrow(linelist)`. Here is our filter statement:
linelist <- linelist %>%
# keep rows where onset is after 1 June 2013 OR where onset is missing and it was a hospital OTHER than Hospital A or B
filter(date_onset > as.Date("2013-06-01") | (is.na(date_onset) & !hospital %in% c("Hospital A", "Hospital B")))
nrow(linelist)## [1] 6019When we re-make the cross-tabulation, we see that Hospitals A & B are removed completely, and the 10 Port Hospital cases from 2012 & 2013 are removed, and all other values are the same - just as we wanted.
table(Hospital = linelist$hospital, # hospital name
YearOnset = lubridate::year(linelist$date_onset), # year of date_onset
useNA = "always") # show missing values## YearOnset
## Hospital 2014 2015 <NA>
## Central Hospital 351 99 18
## Military Hospital 676 200 34
## Missing 1117 318 77
## Other 684 177 46
## Port Hospital 1372 347 75
## St. Mark's Maternity Hospital (SMMH) 322 93 13
## <NA> 0 0 0Multiple statements can be included within one filter command (separated by commas), or you can always pipe to a separate filter() command for clarity.
Note: some readers may notice that it would be easier to just filter by date_hospitalisation because it is 100% complete with no missing values. This is true. But date_onset is used for purposes of demonstrating a complex filter.
Standalone
Filtering can also be done as a stand-alone command (not part of a pipe chain). Like other dplyr verbs, in this case the first argument must be the dataset itself.
# dataframe <- filter(dataframe, condition(s) for rows to keep)
linelist <- filter(linelist, !is.na(case_id))You can also use base R to subset using square brackets which reflect the [rows, columns] that you want to retain.
# dataframe <- dataframe[row conditions, column conditions] (blank means keep all)
linelist <- linelist[!is.na(case_id), ]Quickly review records
Often you want to quickly review a few records, for only a few columns. The base R function View() will print a data frame for viewing in your RStudio.
View the linelist in RStudio:
View(linelist)Here are two examples of viewing specific cells (specific rows, and specific columns):
With dplyr functions filter() and select():
Within View(), pipe the dataset to filter() to keep certain rows, and then to select() to keep certain columns. For example, to review onset and hospitalization dates of 3 specific cases:
View(linelist %>%
filter(case_id %in% c("11f8ea", "76b97a", "47a5f5")) %>%
select(date_onset, date_hospitalisation))You can achieve the same with base R syntax, using brackets [ ] to subset you want to see.
View(linelist[linelist$case_id %in% c("11f8ea", "76b97a", "47a5f5"), c("date_onset", "date_hospitalisation")])Add to pipe chain
# CLEANING 'PIPE' CHAIN (starts with raw data and pipes it through cleaning steps)
##################################################################################
# begin cleaning pipe chain
###########################
linelist <- linelist_raw %>%
# standardize column name syntax
janitor::clean_names() %>%
# manually re-name columns
# NEW name # OLD name
rename(date_infection = infection_date,
date_hospitalisation = hosp_date,
date_outcome = date_of_outcome) %>%
# remove column
select(-c(row_num, merged_header, x28)) %>%
# de-duplicate
distinct() %>%
# add column
mutate(bmi = wt_kg / (ht_cm/100)^2) %>%
# convert class of columns
mutate(across(contains("date"), as.Date),
generation = as.numeric(generation),
age = as.numeric(age)) %>%
# add column: delay to hospitalisation
mutate(days_onset_hosp = as.numeric(date_hospitalisation - date_onset)) %>%
# clean values of hospital column
mutate(hospital = recode(hospital,
# OLD = NEW
"Mitylira Hopital" = "Military Hospital",
"Mitylira Hospital" = "Military Hospital",
"Military Hopital" = "Military Hospital",
"Port Hopital" = "Port Hospital",
"Central Hopital" = "Central Hospital",
"other" = "Other",
"St. Marks Maternity Hopital (SMMH)" = "St. Mark's Maternity Hospital (SMMH)"
)) %>%
mutate(hospital = replace_na(hospital, "Missing")) %>%
# create age_years column (from age and age_unit)
mutate(age_years = case_when(
age_unit == "years" ~ age,
age_unit == "months" ~ age/12,
is.na(age_unit) ~ age,
TRUE ~ NA_real_)) %>%
mutate(
# age categories: custom
age_cat = epikit::age_categories(age_years, breakers = c(0, 5, 10, 15, 20, 30, 50, 70)),
# age categories: 0 to 85 by 5s
age_cat5 = epikit::age_categories(age_years, breakers = seq(0, 85, 5))) %>%
# ABOVE ARE UPSTREAM CLEANING STEPS ALREADY DISCUSSED
###################################################
filter(
# keep only rows where case_id is not missing
!is.na(case_id),
# also filter to keep only the second outbreak
date_onset > as.Date("2013-06-01") | (is.na(date_onset) & !hospital %in% c("Hospital A", "Hospital B")))8.12 Row-wise calculations
If you want to perform a calculation within a row, you can use rowwise() from dplyr. See this online vignette on row-wise calculations.
For example, this code applies rowwise() and then creates a new column that sums the number of the specified symptom columns that have value “yes”, for each row in the linelist. The columns are specified within sum() by name within a vector c(). rowwise() is essentially a special kind of group_by(), so it is best to use ungroup() when you are done (page on Grouping data).
linelist %>%
rowwise() %>%
mutate(num_symptoms = sum(c(fever, chills, cough, aches, vomit) == "yes")) %>%
ungroup() %>%
select(fever, chills, cough, aches, vomit, num_symptoms) # for display## # A tibble: 5,888 x 6
## fever chills cough aches vomit num_symptoms
## <chr> <chr> <chr> <chr> <chr> <int>
## 1 no no yes no yes 2
## 2 <NA> <NA> <NA> <NA> <NA> NA
## 3 <NA> <NA> <NA> <NA> <NA> NA
## 4 no no no no no 0
## 5 no no yes no yes 2
## 6 no no yes no yes 2
## 7 <NA> <NA> <NA> <NA> <NA> NA
## 8 no no yes no yes 2
## 9 no no yes no yes 2
## 10 no no yes no no 1
## # ... with 5,878 more rowsAs you specify the column to evaluate, you may want to use the “tidyselect” helper functions described in the select() section of this page. You just have to make one adjustment (because you are not using them within a dplyr function like select() or summarise()).
Put the column-specification criteria within the dplyr function c_across(). This is because c_across (documentation) is designed to work with rowwise() specifically. For example, the following code:
- Applies
rowwise()so the following operation (sum()) is applied within each row (not summing entire columns)
- Creates new column
num_NA_dates, defined for each row as the number of columns (with name containing “date”) for whichis.na()evaluated to TRUE (they are missing data).
-
ungroup()to remove the effects ofrowwise()for subsequent steps
linelist %>%
rowwise() %>%
mutate(num_NA_dates = sum(is.na(c_across(contains("date"))))) %>%
ungroup() %>%
select(num_NA_dates, contains("date")) # for display## # A tibble: 5,888 x 5
## num_NA_dates date_infection date_onset date_hospitalisation date_outcome
## <int> <date> <date> <date> <date>
## 1 1 2014-05-08 2014-05-13 2014-05-15 NA
## 2 1 NA 2014-05-13 2014-05-14 2014-05-18
## 3 1 NA 2014-05-16 2014-05-18 2014-05-30
## 4 1 2014-05-04 2014-05-18 2014-05-20 NA
## 5 0 2014-05-18 2014-05-21 2014-05-22 2014-05-29
## 6 0 2014-05-03 2014-05-22 2014-05-23 2014-05-24
## 7 0 2014-05-22 2014-05-27 2014-05-29 2014-06-01
## 8 0 2014-05-28 2014-06-02 2014-06-03 2014-06-07
## 9 1 NA 2014-06-05 2014-06-06 2014-06-18
## 10 1 NA 2014-06-05 2014-06-07 2014-06-09
## # ... with 5,878 more rowsYou could also provide other functions, such as max() to get the latest or most recent date for each row:
linelist %>%
rowwise() %>%
mutate(latest_date = max(c_across(contains("date")), na.rm=T)) %>%
ungroup() %>%
select(latest_date, contains("date")) # for display## # A tibble: 5,888 x 5
## latest_date date_infection date_onset date_hospitalisation date_outcome
## <date> <date> <date> <date> <date>
## 1 2014-05-15 2014-05-08 2014-05-13 2014-05-15 NA
## 2 2014-05-18 NA 2014-05-13 2014-05-14 2014-05-18
## 3 2014-05-30 NA 2014-05-16 2014-05-18 2014-05-30
## 4 2014-05-20 2014-05-04 2014-05-18 2014-05-20 NA
## 5 2014-05-29 2014-05-18 2014-05-21 2014-05-22 2014-05-29
## 6 2014-05-24 2014-05-03 2014-05-22 2014-05-23 2014-05-24
## 7 2014-06-01 2014-05-22 2014-05-27 2014-05-29 2014-06-01
## 8 2014-06-07 2014-05-28 2014-06-02 2014-06-03 2014-06-07
## 9 2014-06-18 NA 2014-06-05 2014-06-06 2014-06-18
## 10 2014-06-09 NA 2014-06-05 2014-06-07 2014-06-09
## # ... with 5,878 more rows8.13 Arrange and sort
Use the dplyr function arrange() to sort or order the rows by column values.
Simple list the columns in the order they should be sorted on. Specify .by_group = TRUE if you want the sorting to to first occur by any groupings applied to the data (see page on Grouping data).
By default, column will be sorted in “ascending” order (which applies to numeric and also to character columns). You can sort a variable in “descending” order by wrapping it with desc().
Sorting data with arrange() is particularly useful when making Tables for presentation, using slice() to take the “top” rows per group, or setting factor level order by order of appearance.
For example, to sort the our linelist rows by hospital, then by date_onset in descending order, we would use:
linelist %>%
arrange(hospital, desc(date_onset))